Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model
- PMID: 27013233
- PMCID: PMC4826400
- DOI: 10.1101/gad.276428.115
Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model
Abstract
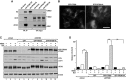
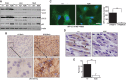
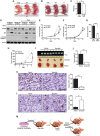
Autophagy is an evolutionarily conserved cellular process controlled through a set of essential autophagy genes (Atgs). However, there is increasing evidence that most, if not all, Atgs also possess functions independent of their requirement in canonical autophagy, making it difficult to distinguish the contributions of autophagy-dependent or -independent functions of a particular Atg to various biological processes. To distinguish these functions for FIP200 (FAK family-interacting protein of 200 kDa), an Atg in autophagy induction, we examined FIP200 interaction with its autophagy partner, Atg13. We found that residues 582-585 (LQFL) in FIP200 are required for interaction with Atg13, and mutation of these residues to AAAA (designated the FIP200-4A mutant) abolished its canonical autophagy function in vitro. Furthermore, we created a FIP200-4A mutant knock-in mouse model and found that specifically blocking FIP200 interaction with Atg13 abolishes autophagy in vivo, providing direct support for the essential role of the ULK1/Atg13/FIP200/Atg101 complex in the process beyond previous studies relying on the complete knockout of individual components. Analysis of the new mouse model showed that nonautophagic functions of FIP200 are sufficient to fully support embryogenesis by maintaining a protective role in TNFα-induced apoptosis. However, FIP200-mediated canonical autophagy is required to support neonatal survival and tumor cell growth. These studies provide the first genetic evidence linking an Atg's autophagy and nonautophagic functions to different biological processes in vivo.
Keywords: FIP200; autophagy; embryogenesis; knock-in mutant mouse; tumor growth.
© 2016 Chen et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Abbi S, Guan JL. 2002. Focal adhesion kinase: protein interactions and cellular functions. Histol Histopathol 17: 1163–1171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
