Importance of a Potential Protein Kinase A Phosphorylation Site of Na+,K+-ATPase and Its Interaction Network for Na+ Binding
- PMID: 27013656
- PMCID: PMC4865937
- DOI: 10.1074/jbc.M115.701201
Importance of a Potential Protein Kinase A Phosphorylation Site of Na+,K+-ATPase and Its Interaction Network for Na+ Binding
Abstract
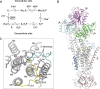
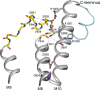
The molecular mechanism underlying PKA-mediated regulation of Na(+),K(+)-ATPase was explored in mutagenesis studies of the potential PKA site at Ser-938 and surrounding charged residues. The phosphomimetic mutations S938D/E interfered with Na(+) binding from the intracellular side of the membrane, whereas Na(+) binding from the extracellular side was unaffected. The reduction of Na(+) affinity is within the range expected for physiological regulation of the intracellular Na(+) concentration, thus supporting the hypothesis that PKA-mediated phosphorylation of Ser-938 regulates Na(+),K(+)-ATPase activity in vivo Ser-938 is located in the intracellular loop between transmembrane segments M8 and M9. An extended bonding network connects this loop with M10, the C terminus, and the Na(+) binding region. Charged residues Asp-997, Glu-998, Arg-1000, and Lys-1001 in M10, participating in this bonding network, are crucial to Na(+) interaction. Replacement of Arg-1005, also located in the vicinity of Ser-938, with alanine, lysine, methionine, or serine resulted in wild type-like Na(+) and K(+) affinities and catalytic turnover rate. However, when combined with the phosphomimetic mutation S938E only lysine substitution of Arg-1005 was compatible with Na(+),K(+)-ATPase function, and the Na(+) affinity of this double mutant was reduced even more than in single mutant S938E. This result indicates that the positive side chain of Arg-1005 or the lysine substituent plays a mechanistic role as interaction partner of phosphorylated Ser-938, transducing the phosphorylation signal into a reduced affinity of Na(+) site III. Electrostatic interaction of Glu-998 is of minor importance for the reduction of Na(+) affinity by phosphomimetic S938E as revealed by combining S938E with E998A.
Keywords: Na+/K+-ATPase; P-type ATPase; PKA site; membrane transport; post-translational regulation; protein kinase A (PKA); site-directed mutagenesis; sodium binding; sodium transport.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Post R. L., Hegyvary C., and Kume S. (1972) Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 247, 6530–6540 - PubMed
-
- Albers R. W. (1967) Biochemical aspects of active transport. Annu. Rev. Biochem. 36, 727–756 - PubMed
-
- Kaplan J. H. (2002) Biochemistry of Na,K-ATPase. Annu. Rev. Biochem. 71, 511–535 - PubMed
-
- Garty H., and Karlish S. J. (2006) Role of FXYD proteins in ion transport. Annu. Rev. Physiol. 68, 431–459 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

