The Fragment 1 Region of Prothrombin Facilitates the Favored Binding of Fragment 12 to Zymogen and Enforces Zymogen-like Character in the Proteinase
- PMID: 27013660
- PMCID: PMC4900261
- DOI: 10.1074/jbc.M116.723072
The Fragment 1 Region of Prothrombin Facilitates the Favored Binding of Fragment 12 to Zymogen and Enforces Zymogen-like Character in the Proteinase
Abstract
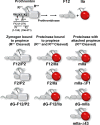
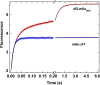
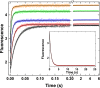
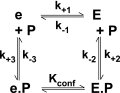
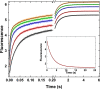
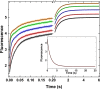
Thrombin is produced from the C-terminal half of prothrombin following its proteolytic activation. The N-terminal half, released as the propiece Fragment 12 (F12), is composed of an N-terminal γ-carboxyglutamate domain (Gla) followed by two kringles (K1 and K2). The propiece plays essential roles in regulating prothrombin activation and proteinase function. The latter results from the ability of F12 to reversibly bind to the (pro)catalytic domain through K2 with high affinity and highly favorable thermodynamic constants when it is a zymogen in comparison to proteinase. Such discrimination is lost for K2 binding after proteolytic removal of the N-terminal Gla-K1 region of F12. The Ca(2+)-stabilized structure of the Gla domain is not required for F12 to bind the zymogen form more favorably. Enhanced binding to zymogen versus proteinase correlates with the ability of the propiece to enforce zymogen-like character in the proteinase. This is evident in variants of meizothrombin, an intermediate of prothrombin activation that contains the propiece covalently attached. This phenomenon is also independent of the Gla domain. Thus, the presence of K1 in covalent linkage with K2 in the propiece governs the ability of K2 to bind the (pro)catalytic domain in favor of zymogen, thereby enforcing zymogen-like character in the proteinase.
Keywords: allosteric regulation; prothrombin; serine protease; thermodynamics; thrombin.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
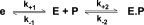


References
-
- Mann K. G., Jenny R. J., and Krishnaswamy S. (1988) Cofactor proteins in the assembly and expression of blood clotting enzyme complexes. Annu. Rev. Biochem. 57, 915–956 - PubMed
-
- Mann K. G., Nesheim M. E., Church W. R., Haley P., and Krishnaswamy S. (1990) Surface-dependent reactions of the vitamin K-dependent enzyme complexes. Blood 76, 1–16 - PubMed
-
- Mann K. G. (2003) Thrombin formation. Chest 124, 4S–10S - PubMed
-
- Jenny N. S., Lundblad R. L., and Mann K. G. (2006) Thrombin. In Hemostasis and Thrombosis. Basic Principles and Clinical Practice (Colman R. W., Marder V. J., Clowes A. J., George J. N., and Goldhaber S. Z., eds.) pp 193–213, Lippincott Williams & Wilkins, Philadelphia
-
- Esmon C. T. (2003) The protein C pathway. Chest 124, 26S–32S - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

