DistAMo: A Web-Based Tool to Characterize DNA-Motif Distribution on Bacterial Chromosomes
- PMID: 27014208
- PMCID: PMC4786541
- DOI: 10.3389/fmicb.2016.00283
DistAMo: A Web-Based Tool to Characterize DNA-Motif Distribution on Bacterial Chromosomes
Abstract
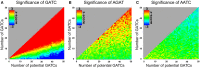
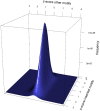
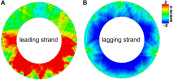
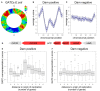
Short DNA motifs are involved in a multitude of functions such as for example chromosome segregation, DNA replication or mismatch repair. Distribution of such motifs is often not random and the specific chromosomal pattern relates to the respective motif function. Computational approaches which quantitatively assess such chromosomal motif patterns are necessary. Here we present a new computer tool DistAMo (Distribution Analysis of DNA Motifs). The algorithm uses codon redundancy to calculate the relative abundance of short DNA motifs from single genes to entire chromosomes. Comparative genomics analyses of the GATC-motif distribution in γ-proteobacterial genomes using DistAMo revealed that (i) genes beside the replication origin are enriched in GATCs, (ii) genome-wide GATC distribution follows a distinct pattern, and (iii) genes involved in DNA replication and repair are enriched in GATCs. These features are specific for bacterial chromosomes encoding a Dam methyltransferase. The new software is available as a stand-alone or as an easy-to-use web-based server version at http://www.computational.bio.uni-giessen.de/distamo.
Keywords: DNA replication; Escherichia coli; algorithm; bacteria; bioinformatics; chromosome maintenance; computational biology.
Figures



Similar articles
-
Arrangement of Dam methylation sites (GATC) in the Escherichia coli chromosome.Nucleic Acids Res. 1988 Oct 25;16(20):9821-38. doi: 10.1093/nar/16.20.9821. Nucleic Acids Res. 1988. PMID: 3054812 Free PMC article.
-
DomainSieve: a protein domain-based screen that led to the identification of dam-associated genes with potential link to DNA maintenance.Bioinformatics. 2006 Aug 15;22(16):1935-41. doi: 10.1093/bioinformatics/btl336. Epub 2006 Jun 20. Bioinformatics. 2006. PMID: 16787973
-
GATC motifs may alter the conformation of DNA depending on sequence context and N6-adenine methylation status: possible implications for DNA-protein recognition.Mol Gen Genet. 1998 Jun;258(5):488-93. doi: 10.1007/s004380050759. Mol Gen Genet. 1998. PMID: 9669330
-
The great GATC: DNA methylation in E. coli.Trends Genet. 1989 May;5(5):139-43. doi: 10.1016/0168-9525(89)90054-1. Trends Genet. 1989. PMID: 2667217 Review.
-
[The DNA-methylation state regulates virulence and stress response of Salmonella].C R Biol. 2008 Sep;331(9):648-54. doi: 10.1016/j.crvi.2008.06.002. Epub 2008 Jul 2. C R Biol. 2008. PMID: 18722983 Review. French.
Cited by
-
The complete methylome of an entomopathogenic bacterium reveals the existence of loci with unmethylated Adenines.Sci Rep. 2018 Aug 14;8(1):12091. doi: 10.1038/s41598-018-30620-5. Sci Rep. 2018. PMID: 30108278 Free PMC article.
-
The Helicobacter pylori orphan ATTAAT-specific methyltransferase M.Hpy99XIX plays a central role in the coordinated regulation of genes involved in iron metabolism.mBio. 2025 Jul 9;16(7):e0120925. doi: 10.1128/mbio.01209-25. Epub 2025 Jun 9. mBio. 2025. PMID: 40488528 Free PMC article.
-
SMRT Sequencing of Paramecium Bursaria Chlorella Virus-1 Reveals Diverse Methylation Stability in Adenines Targeted by Restriction Modification Systems.Front Microbiol. 2020 May 19;11:887. doi: 10.3389/fmicb.2020.00887. eCollection 2020. Front Microbiol. 2020. PMID: 32508769 Free PMC article.
-
Methyltransferase DnmA is responsible for genome-wide N6-methyladenosine modifications at non-palindromic recognition sites in Bacillus subtilis.Nucleic Acids Res. 2020 Jun 4;48(10):5332-5348. doi: 10.1093/nar/gkaa266. Nucleic Acids Res. 2020. PMID: 32324221 Free PMC article.
-
Genome-wide methylome analysis of two strains belonging to the hypervirulent Neisseria meningitidis serogroup W ST-11 clonal complex.Sci Rep. 2021 Mar 18;11(1):6239. doi: 10.1038/s41598-021-85266-7. Sci Rep. 2021. PMID: 33737546 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

