Novel, Deep-Branching Heterotrophic Bacterial Populations Recovered from Thermal Spring Metagenomes
- PMID: 27014227
- PMCID: PMC4791363
- DOI: 10.3389/fmicb.2016.00304
Novel, Deep-Branching Heterotrophic Bacterial Populations Recovered from Thermal Spring Metagenomes
Abstract
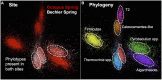
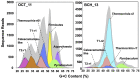
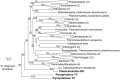
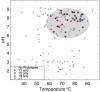
Thermal spring ecosystems are a valuable resource for the discovery of novel hyperthermophilic Bacteria and Archaea, and harbor deeply-branching lineages that provide insight regarding the nature of early microbial life. We characterized bacterial populations in two circumneutral (pH ~8) Yellowstone National Park thermal (T ~80°C) spring filamentous "streamer" communities using random metagenomic DNA sequence to investigate the metabolic potential of these novel populations. Four de novo assemblies representing three abundant, deeply-branching bacterial phylotypes were recovered. Analysis of conserved phylogenetic marker genes indicated that two of the phylotypes represent separate groups of an uncharacterized phylum (for which we propose the candidate phylum name "Pyropristinus"). The third new phylotype falls within the proposed Calescamantes phylum. Metabolic reconstructions of the "Pyropristinus" and Calescamantes populations showed that these organisms appear to be chemoorganoheterotrophs and have the genomic potential for aerobic respiration and oxidative phosphorylation via archaeal-like V-type, and bacterial F-type ATPases, respectively. A survey of similar phylotypes (>97% nt identity) within 16S rRNA gene datasets suggest that the newly described organisms are restricted to terrestrial thermal springs ranging from 70 to 90°C and pH values of ~7-9. The characterization of these lineages is important for understanding the diversity of deeply-branching bacterial phyla, and their functional role in high-temperature circumneutral "streamer" communities.
Keywords: Aquificales; Calescamantes; Pyropristinus; Thermotogae; Yellowstone National Park; hot springs; hyperthermophiles.
Figures




Similar articles
-
Single-Cell-Genomics-Facilitated Read Binning of Candidate Phylum EM19 Genomes from Geothermal Spring Metagenomes.Appl Environ Microbiol. 2015 Dec 4;82(4):992-1003. doi: 10.1128/AEM.03140-15. Print 2016 Feb 15. Appl Environ Microbiol. 2015. PMID: 26637598 Free PMC article.
-
Microbial diversity at 83 degrees C in Calcite Springs, Yellowstone National Park: another environment where the Aquificales and "Korarchaeota" coexist.Extremophiles. 2000 Feb;4(1):61-7. doi: 10.1007/s007920050008. Extremophiles. 2000. PMID: 10741838
-
Spatial and temporal variability of biomarkers and microbial diversity reveal metabolic and community flexibility in Streamer Biofilm Communities in the Lower Geyser Basin, Yellowstone National Park.Geobiology. 2013 Nov;11(6):549-69. doi: 10.1111/gbi.12051. Epub 2013 Aug 28. Geobiology. 2013. PMID: 23981055
-
Uncultivated thermophiles: current status and spotlight on 'Aigarchaeota'.Curr Opin Microbiol. 2015 Jun;25:136-45. doi: 10.1016/j.mib.2015.06.008. Epub 2015 Jun 23. Curr Opin Microbiol. 2015. PMID: 26113243 Review.
-
Assessing the global phylum level diversity within the bacterial domain: A review.J Adv Res. 2015 May;6(3):269-82. doi: 10.1016/j.jare.2014.10.005. Epub 2014 Nov 4. J Adv Res. 2015. PMID: 26257925 Free PMC article. Review.
Cited by
-
An active microbiome in Old Faithful geyser.PNAS Nexus. 2023 Mar 4;2(3):pgad066. doi: 10.1093/pnasnexus/pgad066. eCollection 2023 Mar. PNAS Nexus. 2023. PMID: 37007711 Free PMC article.
-
Diversity and Distribution of a Novel Genus of Hyperthermophilic Aquificae Viruses Encoding a Proof-Reading Family-A DNA Polymerase.Front Microbiol. 2020 Nov 12;11:583361. doi: 10.3389/fmicb.2020.583361. eCollection 2020. Front Microbiol. 2020. PMID: 33281778 Free PMC article.
-
Exploring the taxonomical and functional profile of As Burgas hot spring focusing on thermostable β-galactosidases.Sci Rep. 2021 Jan 8;11(1):101. doi: 10.1038/s41598-020-80489-6. Sci Rep. 2021. PMID: 33420292 Free PMC article.
-
Tectonic settings influence the geochemical and microbial diversity of Peru hot springs.Commun Earth Environ. 2023;4(1):112. doi: 10.1038/s43247-023-00787-5. Epub 2023 Apr 11. Commun Earth Environ. 2023. PMID: 38665187 Free PMC article.
-
Mining microbial and metabolic dark matter in extreme environments: a roadmap for harnessing the power of multi-omics data.Adv Biotechnol (Singap). 2024 Aug 5;2(3):26. doi: 10.1007/s44307-024-00034-8. Adv Biotechnol (Singap). 2024. PMID: 39883228 Free PMC article. Review.
References
-
- Baross J. A., Hoffman S. E. (1985). Submarine hydrothermal vents and associated gradient environments as sites for the origin and evolution of life. Origins Life Evol. B. 15, 327–345. 10.1007/BF01808177 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

