Overlapping Yet Response-Specific Transcriptome Alterations Characterize the Nature of Tobacco-Pseudomonas syringae Interactions
- PMID: 27014286
- PMCID: PMC4779890
- DOI: 10.3389/fpls.2016.00251
Overlapping Yet Response-Specific Transcriptome Alterations Characterize the Nature of Tobacco-Pseudomonas syringae Interactions
Abstract
In this study transcriptomic alterations of bacterially induced pattern triggered immunity (PTI) were compared with other types of tobacco-Pseudomonas interactions. In addition, using pharmacological agents we blocked some signal transduction pathways (Ca(2+) influx, kinases, phospholipases, proteasomic protein degradation) to find out how they contribute to gene expression during PTI. PTI is the first defense response of plant cells to microbes, elicited by their widely conserved molecular patterns. Tobacco is an important model of Solanaceae to study resistance responses, including defense mechanisms against bacteria. In spite of these facts the transcription regulation of tobacco genes during different types of plant bacterial interactions is not well-described. In this paper we compared the tobacco transcriptomic alterations in microarray experiments induced by (i) PTI inducer Pseudomonas syringae pv. syringae type III secretion mutant (hrcC) at earlier (6 h post inoculation) and later (48 hpi) stages of defense, (ii) wild type P. syringae (6 hpi) that causes effector triggered immunity (ETI) and cell death (HR), and (iii) disease-causing P. syringae pv. tabaci (6 hpi). Among the different treatments the highest overlap was between the PTI and ETI at 6 hpi, however, there were groups of genes with specifically altered activity for either type of defenses. Instead of quantitative effects of the virulent P. tabaci on PTI-related genes it influenced transcription qualitatively and blocked the expression changes of a special set of genes including ones involved in signal transduction and transcription regulation. P. tabaci specifically activated or repressed other groups of genes seemingly not related to either PTI or ETI. Kinase and phospholipase A inhibitors had highest impacts on the PTI response and effects of these signal inhibitors on transcription greatly overlapped. Remarkable interactions of phospholipase C-related pathways with the proteasomal system were also observable. Genes specifically affected by virulent P. tabaci belonged to various previously identified signaling routes, suggesting that compatible pathogens may modulate diverse signaling pathways of PTI to overcome plant defense.
Keywords: Pseudomonas syringae; compatible interaction; effector triggered immunity (ETI); pattern triggered immunity (PTI); signal transduction; tobacco; transcriptome.
Figures
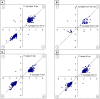

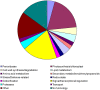

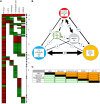
References
-
- Albrecht C., Boutrot F., Segonzac C., Schwessinger B., Gimenez-Ibanez S., Chinchilla D., et al. (2012). Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. U.S.A. 109, 303–308. 10.1073/pnas.1109921108 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

