The Evolutionarily Conserved E3 Ubiquitin Ligase AtCHIP Contributes to Plant Immunity
- PMID: 27014328
- PMCID: PMC4791365
- DOI: 10.3389/fpls.2016.00309
The Evolutionarily Conserved E3 Ubiquitin Ligase AtCHIP Contributes to Plant Immunity
Abstract
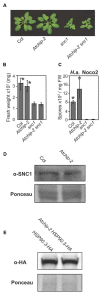
Plants possess a sophisticated immune system to recognize and respond to microbial threats in their environment. The level of immune signaling must be tightly regulated so that immune responses can be quickly activated in the presence of pathogens, while avoiding autoimmunity. HSP90s, along with their diverse array of co-chaperones, forms chaperone complexes that have been shown to play both positive and negative roles in regulating the accumulation of immune receptors and regulators. In this study, we examined the role of AtCHIP, an evolutionarily conserved E3 ligase that was known to interact with chaperones including HSP90s in multicellular organisms including fruit fly, Caenorhabditis elegans, plants and human. Atchip knockout mutants display enhanced disease susceptibility to a virulent oomycete pathogen, and overexpression of AtCHIP causes enhanced disease resistance at low temperature. Although CHIP was reported to target HSP90 for ubiquitination and degradation, accumulation of HSP90.3 was not affected in Atchip plants. In addition, protein accumulation of nucleotide-binding, leucine-rich repeat domain immune receptor (NLR) SNC1 is not altered in Atchip mutant. Thus, while AtCHIP plays a role in immunity, it does not seem to regulate the turnover of HSP90 or SNC1. Further investigation is needed in order to determine the exact mechanism behind AtCHIP's role in regulating plant immune responses.
Keywords: ChIP; E3 lligase; HSP90 heat-shock proteins; SGT1; chaperones; plant immunity.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

