Genetic and life-history traits associated with the distribution of prophages in bacteria
- PMID: 27015004
- PMCID: PMC5113838
- DOI: 10.1038/ismej.2016.47
Genetic and life-history traits associated with the distribution of prophages in bacteria
Abstract
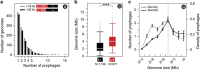
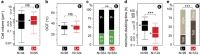
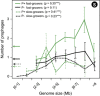
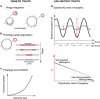
Nearly half of the sequenced bacteria are lysogens and many of their prophages encode adaptive traits. Yet, the variables driving prophage distribution remain undetermined. We identified 2246 prophages in complete bacterial genomes to study the genetic and life-history traits associated with lysogeny. While optimal growth temperatures and average cell volumes were not associated with lysogeny, prophages were more frequent in pathogens and in bacteria with small minimal doubling times. Their frequency also increased with genome size, but only for genomes smaller than 6 Mb. The number of spacers in CRISPR-Cas systems and the frequency of type III systems were anticorrelated with prophage frequency, but lysogens were more likely to encode type I and type II systems. The minimal doubling time was the trait most correlated with lysogeny, followed by genome size and pathogenicity. We propose that bacteria with highly variable growth rates often encounter lower opportunity costs for lysogeny relative to lysis. These results contribute to explain the paucity of temperate phages in certain bacterial clades and of bacterial lysogens in certain environments. They suggest that genetic and life-history traits affect the contributions of temperate phages to bacterial genomes.
Figures

References
-
- Abedon ST. (2008) Bacteriophage Ecology: Population Growth, Evolution, and Impact of Bacterial Viruses. Cambridge University Press: Cambridge, NY, USA.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

