Heterogeneity in Oct4 and Sox2 Targets Biases Cell Fate in 4-Cell Mouse Embryos
- PMID: 27015307
- PMCID: PMC4819611
- DOI: 10.1016/j.cell.2016.01.047
Heterogeneity in Oct4 and Sox2 Targets Biases Cell Fate in 4-Cell Mouse Embryos
Abstract
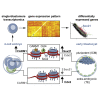
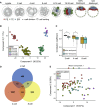
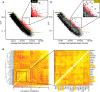
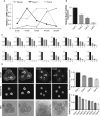
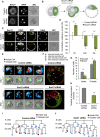
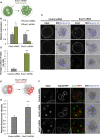
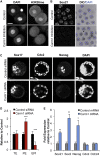
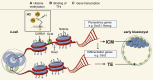
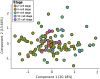
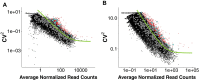
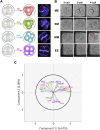
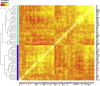
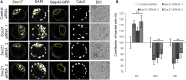
The major and essential objective of pre-implantation development is to establish embryonic and extra-embryonic cell fates. To address when and how this fundamental process is initiated in mammals, we characterize transcriptomes of all individual cells throughout mouse pre-implantation development. This identifies targets of master pluripotency regulators Oct4 and Sox2 as being highly heterogeneously expressed between blastomeres of the 4-cell embryo, with Sox21 showing one of the most heterogeneous expression profiles. Live-cell tracking demonstrates that cells with decreased Sox21 yield more extra-embryonic than pluripotent progeny. Consistently, decreasing Sox21 results in premature upregulation of the differentiation regulator Cdx2, suggesting that Sox21 helps safeguard pluripotency. Furthermore, Sox21 is elevated following increased expression of the histone H3R26-methylase CARM1 and is lowered following CARM1 inhibition, indicating the importance of epigenetic regulation. Therefore, our results indicate that heterogeneous gene expression, as early as the 4-cell stage, initiates cell-fate decisions by modulating the balance of pluripotency and differentiation.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Comment in
-
The Molecular Harbingers of Early Mammalian Embryo Patterning.Cell. 2016 Mar 24;165(1):13-15. doi: 10.1016/j.cell.2016.03.005. Cell. 2016. PMID: 27015301
-
Investigations at the 'Four-Front' of Mammalian Development.Trends Genet. 2016 Aug;32(8):457-458. doi: 10.1016/j.tig.2016.05.005. Epub 2016 May 30. Trends Genet. 2016. PMID: 27256906 Free PMC article.
References
-
- Brennecke P., Anders S., Kim J.K., Kołodziejczyk A.A., Zhang X., Proserpio V., Baying B., Benes V., Teichmann S.A., Marioni J.C., Heisler M.G. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods. 2013;10:1093–1095. - PubMed
-
- Buettner F., Natarajan K.N., Casale F.P., Proserpio V., Scialdone A., Theis F.J., Teichmann S.A., Marioni J.C., Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 2015;33:155–160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

