The Fuzzy Logic of Network Connectivity in Mouse Visual Thalamus
- PMID: 27015312
- PMCID: PMC4808248
- DOI: 10.1016/j.cell.2016.02.033
The Fuzzy Logic of Network Connectivity in Mouse Visual Thalamus
Abstract
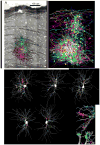
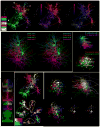
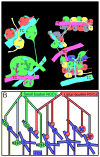
In an attempt to chart parallel sensory streams passing through the visual thalamus, we acquired a 100-trillion-voxel electron microscopy (EM) dataset and identified cohorts of retinal ganglion cell axons (RGCs) that innervated each of a diverse group of postsynaptic thalamocortical neurons (TCs). Tracing branches of these axons revealed the set of TCs innervated by each RGC cohort. Instead of finding separate sensory pathways, we found a single large network that could not be easily subdivided because individual RGCs innervated different kinds of TCs and different kinds of RGCs co-innervated individual TCs. We did find conspicuous network subdivisions organized on the basis of dendritic rather than neuronal properties. This work argues that, in the thalamus, neural circuits are not based on a canonical set of connections between intrinsically different neuronal types but, rather, may arise by experience-based mixing of different kinds of inputs onto individual postsynaptic cells.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Untangling the Web between Eye and Brain.Cell. 2016 Mar 24;165(1):20-21. doi: 10.1016/j.cell.2016.03.010. Cell. 2016. PMID: 27015304 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

