A Potential Structural Switch for Regulating DNA-Binding by TEAD Transcription Factors
- PMID: 27016204
- PMCID: PMC4893915
- DOI: 10.1016/j.jmb.2016.03.008
A Potential Structural Switch for Regulating DNA-Binding by TEAD Transcription Factors
Abstract
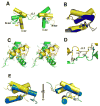
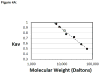
TEA domain (TEAD) transcription factors are essential for the normal development of eukaryotes and are the downstream effectors of the Hippo tumor suppressor pathway. Whereas our earlier work established the three-dimensional structure of the highly conserved DNA-binding domain using solution NMR spectroscopy, the structural basis for regulating the DNA-binding activity remains unknown. Here, we present the X-ray crystallographic structure and activity of a TEAD mutant containing a truncated L1 loop, ΔL1 TEAD DBD. Unexpectedly, the three-dimensional structure of the ΔL1 TEAD DBD reveals a helix-swapped homodimer wherein helix 1 is swapped between monomers. Furthermore, each three-helix bundle in the domain-swapped dimer is a structural homolog of MYB-like domains. Our investigations of the DNA-binding activity reveal that although the formation of the three-helix bundle by the ΔL1 TEAD DBD is sufficient for binding to an isolated M-CAT-like DNA element, multimeric forms are deficient for cooperative binding to tandemly duplicated elements, indicating that the L1 loop contributes to the DNA-binding activity of TEAD. These results suggest that switching between monomeric and domain-swapped forms may regulate DNA selectivity of TEAD proteins.
Keywords: Hippo pathway; TEAD; X-ray crystallography; domain swapping; transcription factor.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures




Similar articles
-
DNA-binding mechanism of the Hippo pathway transcription factor TEAD4.Oncogene. 2017 Jul 27;36(30):4362-4369. doi: 10.1038/onc.2017.24. Epub 2017 Apr 3. Oncogene. 2017. PMID: 28368398
-
Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain.Proc Natl Acad Sci U S A. 2006 Nov 14;103(46):17225-30. doi: 10.1073/pnas.0607171103. Epub 2006 Nov 3. Proc Natl Acad Sci U S A. 2006. PMID: 17085591 Free PMC article.
-
Crystal structure of the human LRH-1 DBD-DNA complex reveals Ftz-F1 domain positioning is required for receptor activity.J Mol Biol. 2005 Dec 16;354(5):1091-102. doi: 10.1016/j.jmb.2005.10.009. Epub 2005 Oct 27. J Mol Biol. 2005. PMID: 16289203
-
The virtuoso of versatility: POU proteins that flex to fit.J Mol Biol. 2000 Oct 6;302(5):1023-39. doi: 10.1006/jmbi.2000.4107. J Mol Biol. 2000. PMID: 11183772 Review.
-
An evolutionary, structural and functional overview of the mammalian TEAD1 and TEAD2 transcription factors.Gene. 2016 Oct 10;591(1):292-303. doi: 10.1016/j.gene.2016.07.028. Epub 2016 Jul 14. Gene. 2016. PMID: 27421669 Free PMC article. Review.
Cited by
-
Exploring trophoblast-specific Tead4 enhancers through chromatin conformation capture assays followed by functional screening.Nucleic Acids Res. 2020 Jan 10;48(1):278-289. doi: 10.1093/nar/gkz1034. Nucleic Acids Res. 2020. PMID: 31777916 Free PMC article.
-
Targeting the Hippo Pathway and Cancer through the TEAD Family of Transcription Factors.Cancers (Basel). 2018 Mar 20;10(3):81. doi: 10.3390/cancers10030081. Cancers (Basel). 2018. PMID: 29558384 Free PMC article. Review.
-
TEAD2 initiates ground-state pluripotency by mediating chromatin looping.EMBO J. 2024 May;43(10):1965-1989. doi: 10.1038/s44318-024-00086-5. Epub 2024 Apr 11. EMBO J. 2024. PMID: 38605224 Free PMC article.
-
DNA-binding mechanism of the Hippo pathway transcription factor TEAD4.Oncogene. 2017 Jul 27;36(30):4362-4369. doi: 10.1038/onc.2017.24. Epub 2017 Apr 3. Oncogene. 2017. PMID: 28368398
-
Biochemical properties of VGLL4 from Homo sapiens and Tgi from Drosophila melanogaster and possible biological implications.Protein Sci. 2021 Sep;30(9):1871-1881. doi: 10.1002/pro.4138. Epub 2021 Jun 8. Protein Sci. 2021. PMID: 34075638 Free PMC article.
References
-
- Halder G, Carroll SB. Binding of the Vestigial co-factor switches the DNA-target selectivity of the Scalloped selector protein. Development. 2001;128:3295–305. - PubMed
-
- Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development and the TEAD family of transcription factors. Dev Genet. 1998;22:43–55. - PubMed
-
- Davidson I, Xiao JH, Rosales R, Staub A, Chambon P. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 1988;54:931–42. - PubMed
-
- Jiang SW, Desai D, Khan S, Eberhardt NL. Cooperative binding of TEF-1 to repeated GGAATG-related consensus elements with restricted spatial separation and orientation. DNA Cell Biol. 2000;19:507–14. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

