Structural Dynamics and Mechanochemical Coupling in DNA Gyrase
- PMID: 27016205
- PMCID: PMC5083069
- DOI: 10.1016/j.jmb.2016.03.016
Structural Dynamics and Mechanochemical Coupling in DNA Gyrase
Abstract
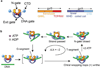
Gyrase is a molecular motor that harnesses the free energy of ATP hydrolysis to perform mechanical work on DNA. The enzyme specifically introduces negative supercoiling in a process that must coordinate fuel consumption with DNA cleavage and religation and with numerous conformational changes in both the protein and DNA components of a large nucleoprotein complex. Here we present a current understanding of mechanochemical coupling in this essential molecular machine, with a focus on recent diverse biophysical approaches that have revealed details of molecular architectures, new conformational intermediates, structural transitions modulated by ATP binding, and the influence of mechanics on motor function. Recent single-molecule assays have also illuminated the reciprocal relationships between supercoiling and transcription, an illustration of mechanical interactions between gyrase and other molecular machines at the heart of chromosomal biology.
Keywords: FRET; magnetic tweezers; molecular motor; single-molecule; topoisomerase.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures




References
-
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. - PubMed
-
- Nollmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie. 2007;89:490–499. - PubMed
-
- Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Quarterly reviews of biophysics. 2008;41:41–101. - PubMed
-
- Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. - PubMed
-
- Bax BD, Chan PF, Eggleston DS, Fosberry A, Gentry DR, Gorrec F, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

