Transcriptome analyses of insect cells to facilitate baculovirus-insect expression
- PMID: 27017378
- PMCID: PMC4853316
- DOI: 10.1007/s13238-016-0260-y
Transcriptome analyses of insect cells to facilitate baculovirus-insect expression
Abstract
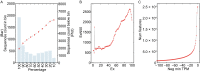
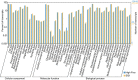
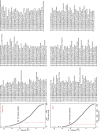
The High Five cell line (BTI-TN-5B1-4) isolated from the cabbage looper, Trichoplusia ni is an insect cell line widely used for baculovirus-mediated recombinant protein expression. Despite its widespread application in industry and academic laboratories, the genomic background of this cell line remains unclear. Here we sequenced the transcriptome of High Five cells and assembled 25,234 transcripts. Codon usage analysis showed that High Five cells have a robust codon usage capacity and therefore suit for expressing proteins of both eukaryotic- and prokaryotic-origin. Genes involved in glycosylation were profiled in our study, providing guidance for engineering glycosylated proteins in the insect cells. We also predicted signal peptides for transcripts with high expression abundance in both High Five and Sf21 cell lines, and these results have important implications for optimizing the expression level of some secretory and membrane proteins.
Keywords: High Five cell line; baculovirus-insect cell system; codon usage; glycosylation; signal peptide.
Figures


References
-
- Cannarozzi GM, Schneider A (eds) (2012) Codon Evolution. Oxford University Press
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

