SMART precision cancer medicine: a FHIR-based app to provide genomic information at the point of care
- PMID: 27018265
- PMCID: PMC6080684
- DOI: 10.1093/jamia/ocw015
SMART precision cancer medicine: a FHIR-based app to provide genomic information at the point of care
Abstract
Background: Precision cancer medicine (PCM) will require ready access to genomic data within the clinical workflow and tools to assist clinical interpretation and enable decisions. Since most electronic health record (EHR) systems do not yet provide such functionality, we developed an EHR-agnostic, clinico-genomic mobile app to demonstrate several features that will be needed for point-of-care conversations.
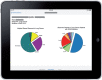
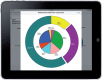
Methods: Our prototype, called Substitutable Medical Applications and Reusable Technology (SMART)® PCM, visualizes genomic information in real time, comparing a patient's diagnosis-specific somatic gene mutations detected by PCR-based hotspot testing to a population-level set of comparable data. The initial prototype works for patient specimens with 0 or 1 detected mutation. Genomics extensions were created for the Health Level Seven® Fast Healthcare Interoperability Resources (FHIR)® standard; otherwise, the prototype is a normal SMART on FHIR app.
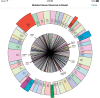
Results: The PCM prototype can rapidly present a visualization that compares a patient's somatic genomic alterations against a distribution built from more than 3000 patients, along with context-specific links to external knowledge bases. Initial evaluation by oncologists provided important feedback about the prototype's strengths and weaknesses. We added several requested enhancements and successfully demonstrated the app at the inaugural American Society of Clinical Oncology Interoperability Demonstration; we have also begun to expand visualization capabilities to include cancer specimens with multiple mutations.
Discussion: PCM is open-source software for clinicians to present the individual patient within the population-level spectrum of cancer somatic mutations. The app can be implemented on any SMART on FHIR-enabled EHRs, and future versions of PCM should be able to evolve in parallel with external knowledge bases.
Keywords: electronic health records; genomics; health information management; information science; mobile health; neoplasms.
© The Author 2016. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

Similar articles
-
Opening the Duke electronic health record to apps: Implementing SMART on FHIR.Int J Med Inform. 2017 Mar;99:1-10. doi: 10.1016/j.ijmedinf.2016.12.005. Epub 2016 Dec 12. Int J Med Inform. 2017. PMID: 28118917
-
SMART on FHIR: a standards-based, interoperable apps platform for electronic health records.J Am Med Inform Assoc. 2016 Sep;23(5):899-908. doi: 10.1093/jamia/ocv189. Epub 2016 Feb 17. J Am Med Inform Assoc. 2016. PMID: 26911829 Free PMC article.
-
SMART-on-FHIR implemented over i2b2.J Am Med Inform Assoc. 2017 Mar 1;24(2):398-402. doi: 10.1093/jamia/ocw079. J Am Med Inform Assoc. 2017. PMID: 27274012 Free PMC article.
-
Patient-Centered Radiology with FHIR: an Introduction to the Use of FHIR to Offer Radiology a Clinically Integrated Platform.J Digit Imaging. 2018 Jun;31(3):327-333. doi: 10.1007/s10278-018-0087-6. J Digit Imaging. 2018. PMID: 29725963 Free PMC article. Review.
-
Learning HL7 FHIR Using the HAPI FHIR Server and Its Use in Medical Imaging with the SIIM Dataset.J Digit Imaging. 2018 Jun;31(3):334-340. doi: 10.1007/s10278-018-0090-y. J Digit Imaging. 2018. PMID: 29725959 Free PMC article. Review.
Cited by
-
Fast Healthcare Interoperability Resources (FHIR)-Based Quality Information Exchange for Clinical Next-Generation Sequencing Genomic Testing: Implementation Study.J Med Internet Res. 2021 Apr 28;23(4):e26261. doi: 10.2196/26261. J Med Internet Res. 2021. PMID: 33908889 Free PMC article.
-
Genetic and Genomic Competencies for Nursing Informatics Internationally.Stud Health Technol Inform. 2017;232:152-164. Stud Health Technol Inform. 2017. PMID: 28106593 Free PMC article.
-
MatchMiner: an open-source platform for cancer precision medicine.NPJ Precis Oncol. 2022 Oct 6;6(1):69. doi: 10.1038/s41698-022-00312-5. NPJ Precis Oncol. 2022. PMID: 36202909 Free PMC article.
-
Molecularly-Guided Cancer Clinical Trial Matching using FHIR and HL7 Clinical Quality Language: A Proof of Concept.AMIA Annu Symp Proc. 2025 May 22;2024:359-367. eCollection 2024. AMIA Annu Symp Proc. 2025. PMID: 40417499 Free PMC article.
-
SMART Cancer Navigator: A Framework for Implementing ASCO Workshop Recommendations to Enable Precision Cancer Medicine.JCO Precis Oncol. 2018;2018:PO.17.00292. doi: 10.1200/PO.17.00292. Epub 2018 May 1. JCO Precis Oncol. 2018. PMID: 30238071 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

