Quantification of cellular and nuclear uptake rates of polymeric gene delivery nanoparticles and DNA plasmids via flow cytometry
- PMID: 27019146
- PMCID: PMC5061650
- DOI: 10.1016/j.actbio.2016.03.036
Quantification of cellular and nuclear uptake rates of polymeric gene delivery nanoparticles and DNA plasmids via flow cytometry
Abstract
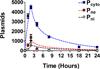
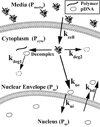
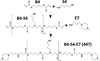
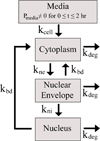
Non-viral, biomaterial-mediated gene delivery has the potential to treat many diseases, but is limited by low efficacy. Elucidating the bottlenecks of plasmid mass transfer can enable an improved understanding of biomaterial structure-function relationships, leading to next-generation rationally designed non-viral gene delivery vectors. As proof of principle, we transfected human primary glioblastoma cells using a poly(beta-amino ester) complexed with eGFP plasmid DNA. The polyplexes transfected 70.6±0.6% of the cells with 101±3% viability. The amount of DNA within the cytoplasm, nuclear envelope, and nuclei was assessed at multiple time points using fluorescent dye conjugated plasmid up to 24h post-transfection using a quantitative multi-well plate-based flow cytometry assay. Conversion to plasmid counts and degradation kinetics were accounted for via quantitative PCR (plasmid degradation rate constants were determined to be 0.62h(-1) and 0.084h(-1) for fast and slow phases respectively). Quantitative cellular uptake, nuclear association, and nuclear uptake rate constants were determined by using a four-compartment first order mass-action model. The rate limiting step for these poly(beta-amino ester)/DNA polyplex nanoparticles was determined to be cellular uptake (7.5×10(-4)h(-1)) and only 0.1% of the added dose was taken up by the human brain cancer cells, whereas 12% of internalized DNA successfully entered the nucleus (the rate of nuclear internalization of nuclear associated plasmid was 1.1h(-1)). We describe an efficient new method for assessing cellular and nuclear uptake rates of non-viral gene delivery nanoparticles using flow cytometry to improve understanding and design of polymeric gene delivery nanoparticles.
Statement of significance: In this work, a quantitative high throughput flow cytometry-based assay and computational modeling approach was developed for assessing cellular and nuclear uptake rates of non-viral gene delivery nanoparticles. This method is significant as it can be used to elucidate structure-function relationships of gene delivery nanoparticles and improve their efficiency. This method was applied to a particular type of biodegradable polymer, a poly(beta-amino ester), that transfected human brain cancer cells with high efficacy and without cytotoxicity. A four-compartment first order mass-action kinetics model was found to model the experimental transport data well without requiring external fitting parameters. Quantitative rate constants were identified for the intracellular transport, including DNA degradation rate from polyplexes, cellular uptake rate, and nuclear uptake rate, with cellular uptake identified as the rate-limiting step.
Keywords: Brain cancer; Computational modeling; Gene delivery; Nanoparticle; Polymer.
Copyright © 2016 Acta Materialia Inc. Published by Elsevier Ltd. All rights reserved.
Figures




Similar articles
-
Synthesis and application of poly(ethylene glycol)-co-poly(β-amino ester) copolymers for small cell lung cancer gene therapy.Acta Biomater. 2016 Sep 1;41:293-301. doi: 10.1016/j.actbio.2016.05.040. Epub 2016 Jun 1. Acta Biomater. 2016. PMID: 27262740 Free PMC article.
-
A high-throughput bioimaging study to assess the impact of chitosan-based nanoparticle degradation on DNA delivery performance.Acta Biomater. 2016 Dec;46:129-140. doi: 10.1016/j.actbio.2016.09.037. Epub 2016 Sep 26. Acta Biomater. 2016. PMID: 27686038
-
Evaluation of polymeric gene delivery nanoparticles by nanoparticle tracking analysis and high-throughput flow cytometry.J Vis Exp. 2013 Mar 1;(73):e50176. doi: 10.3791/50176. J Vis Exp. 2013. PMID: 23486314 Free PMC article.
-
Exploring the role of polymer structure on intracellular nucleic acid delivery via polymeric nanoparticles.J Control Release. 2015 Dec 10;219:488-499. doi: 10.1016/j.jconrel.2015.09.046. Epub 2015 Oct 1. J Control Release. 2015. PMID: 26433125 Free PMC article. Review.
-
Intracellular barriers to non-viral gene transfer.Curr Gene Ther. 2002 May;2(2):183-94. doi: 10.2174/1566523024605609. Curr Gene Ther. 2002. PMID: 12109215 Review.
Cited by
-
Non-viral Delivery of Nucleic Acids: Insight Into Mechanisms of Overcoming Intracellular Barriers.Front Pharmacol. 2018 Aug 21;9:971. doi: 10.3389/fphar.2018.00971. eCollection 2018. Front Pharmacol. 2018. PMID: 30186185 Free PMC article. Review.
-
A Triple-Fluorophore-Labeled Nucleic Acid pH Nanosensor to Investigate Non-viral Gene Delivery.Mol Ther. 2017 Jul 5;25(7):1697-1709. doi: 10.1016/j.ymthe.2017.04.008. Epub 2017 May 4. Mol Ther. 2017. PMID: 28479046 Free PMC article.
-
DNA unchained: two assays to discover and study inhibitors of the DNA clustering function of barrier-to-autointegration factor.Sci Rep. 2020 Jul 23;10(1):12301. doi: 10.1038/s41598-020-69246-x. Sci Rep. 2020. PMID: 32704141 Free PMC article.
-
Nanoparticles for Stem Cell Therapy Bioengineering in Glioma.Front Bioeng Biotechnol. 2020 Dec 7;8:558375. doi: 10.3389/fbioe.2020.558375. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33365304 Free PMC article. Review.
-
Transient Destabilization of Biological Membranes Contributes to the Superior Performance of Star-Shaped PDMAEMA in Delivering pDNA.ACS Omega. 2020 Oct 5;5(41):26640-26654. doi: 10.1021/acsomega.0c03367. eCollection 2020 Oct 20. ACS Omega. 2020. PMID: 33110991 Free PMC article.
References
-
- Alton EW, Middleton PG, Caplen NJ, Smith SN, Steel DM, Munkonge FM, Jeffery PK, Geddes DM, Hart SL, Williamson R, et al. Non-invasive liposome-mediated gene delivery can correct the ion transport defect in cystic fibrosis mutant mice. Nat. Genet. 1993;5:135–142. - PubMed
-
- Nathwani AC, Tuddenham EGD, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, O'Beirne J, Smith K, Pasi J, Glader B, Rustagi P, Ng CYC, Kay MA, Zhou JF, Spence Y, Morton CL, Allay J, Coleman J, Sleep S, Cunningham JM, Srivastava D, Basner-Tschakarjan E, Mingozzi F, High KA, Gray JT, Reiss UM, Nienhuis AW, Davidoff AM. Adenovirus-Associated Virus Vector-Mediated Gene Transfer in Hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

