Associations with rhizosphere bacteria can confer an adaptive advantage to plants
- PMID: 27019743
- PMCID: PMC4806546
- DOI: 10.1038/nplants.2015.51
Associations with rhizosphere bacteria can confer an adaptive advantage to plants
Abstract
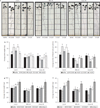
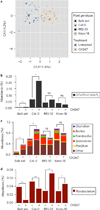
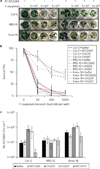
Host-associated microbiomes influence host health. However, it is unclear whether genotypic variations in host organisms influence the microbiome in ways that have adaptive consequences for the host. Here, we show that wild accessions of Arabidopsis thaliana differ in their ability to associate with the root-associated bacterium Pseudomonas fluorescens, with consequences for plant fitness. In a screen of 196 naturally occurring Arabidopsis accessions we identified lines that actively suppress Pseudomonas growth under gnotobiotic conditions. We planted accessions that support disparate levels of fluorescent Pseudomonads in natural soils; 16S ribosomal RNA sequencing revealed that accession-specific differences in the microbial communities were largely limited to a subset of Pseudomonadaceae species. These accession-specific differences in Pseudomonas growth resulted in enhanced or impaired fitness that depended on the host's ability to support Pseudomonas growth, the specific Pseudomonas strains present in the soil and the nature of the stress. We suggest that small host-mediated changes in a microbiome can have large effects on host health.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

