Functional Assessment of Clubfoot Associated HOXA9, TPM1, and TPM2 Variants Suggests a Potential Gene Regulation Mechanism
- PMID: 27020427
- PMCID: PMC4887369
- DOI: 10.1007/s11999-016-4788-1
Functional Assessment of Clubfoot Associated HOXA9, TPM1, and TPM2 Variants Suggests a Potential Gene Regulation Mechanism
Abstract
Background: Isolated nonsyndromic clubfoot is a common birth defect affecting 135,000 newborns worldwide each year. Although treatment has improved, substantial long-term morbidity persists. Genetic causes have been implicated in family-based studies but the genetic changes have eluded identification. Previously, using a candidate gene approach in our family-based dataset, we identified associations between clubfoot and four single nucleotide polymorphisms (SNPs) located in potential regulatory regions of genes involved in muscle development and patterning (HOXA9) and muscle function (TPM1 and TPM2) were identified.
Questions/purposes: Four SNPs, rs3801776/HOXA9, rs4075583/TPM1, rs2025126/TPM2, and rs2145925/TPM2, located in potential regulatory regions, were evaluated to determine whether they altered promoter activity.
Methods: Electrophoretic mobility shift assays were performed on these four SNPs to identify allele-specific DNA-protein interactions. SNPs showing differential banding patterns were assessed for effect on promoter activity by luciferase assay. Undifferentiated (for HOXA9) and differentiated (for TPM1 and TPM2) mouse cells were used in functional assays as a proxy for the in vivo developmental stage.
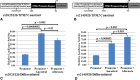
Results: Functional analyses showed that the ancestral alleles of rs3801776/HOXA9, rs4075583/TPM1, and rs2025126/TPM2 and the alternate allele of rs2145925/TPM2 created allele-specific nuclear protein interactions and caused higher promoter activity. Interestingly, while rs4075583/TPM1 showed an allele-specific nuclear protein interaction, an effect on promoter activity was observed only when rs4075583/TPM1 was expressed in the 1.7kb haplotype construct.
Conclusion: Our results show that associated promoter variants in HOXA9, TPM1, and TPM2, alter promoter expression suggesting that they have a functional role. Moreover and importantly, we show that alterations in promoter activity may be observed only in the context of the genomic architecture. Therefore, future studies focusing on proteins binding to these regulatory SNPs may provide important key insights into gene regulation in clubfoot.
Clinical relevance: Identifying the genetic risk signature for clubfoot is important to provide accurate genetic counseling for at-risk families, for development of prevention programs and new treatments.
Figures



References
-
- Alvarado DM, Aferol H, McCall K, Huang JB, Techy M, Buchan J, Cady J, Gonzales PR, Dobbs MB, Gurnett CA. Familial isolated clubfoot is associated with recurrent chromosome 17q23.1q23.2 microduplications containing TBX4. Am J Hum Genet. 2010;87:154–160. doi: 10.1016/j.ajhg.2010.06.010. - DOI - PMC - PubMed
-
- Beals RK. Club foot in the Maori: a genetic study of 50 kindreds. N Z Med J. 1978;88:144–146. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

