Local Pheromone Release from Dynamic Polarity Sites Underlies Cell-Cell Pairing during Yeast Mating
- PMID: 27020743
- PMCID: PMC4846541
- DOI: 10.1016/j.cub.2016.02.064
Local Pheromone Release from Dynamic Polarity Sites Underlies Cell-Cell Pairing during Yeast Mating
Abstract
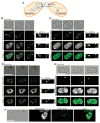
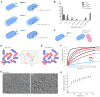
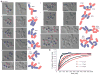
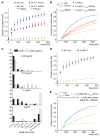
Cell pairing is central for many processes, including immune defense, neuronal connection, hyphal fusion, and sexual reproduction. How does a cell orient toward a partner, especially when faced with multiple choices? Fission yeast Schizosaccharomyces pombe P and M cells, which respectively express P and M factor pheromones [1, 2], pair during the mating process induced by nitrogen starvation. Engagement of pheromone receptors Map3 and Mam2 [3, 4] with their cognate pheromone ligands leads to activation of the Gα protein Gpa1 to signal sexual differentiation [3, 5, 6]. Prior to cell pairing, the Cdc42 GTPase, a central regulator of cell polarization, forms dynamic zones of activity at the cell periphery at distinct locations over time [7]. Here we show that Cdc42-GTP polarization sites contain the M factor transporter Mam1, the general secretion machinery, which underlies P factor secretion, and Gpa1, suggesting that these are sub-cellular zones of pheromone secretion and signaling. Zone lifetimes scale with pheromone concentration. Computational simulations of pair formation through a fluctuating zone show that the combination of local pheromone release and sensing, short pheromone decay length, and pheromone-dependent zone stabilization leads to efficient pair formation. Consistently, pairing efficiency is reduced in the absence of the P factor protease. Similarly, zone stabilization at reduced pheromone levels, which occurs in the absence of the predicted GTPase-activating protein for Ras, leads to reduction in pairing efficiency. We propose that efficient cell pairing relies on fluctuating local signal emission and perception, which become locked into place through stimulation.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
Comment in
-
Gradient Sensing: Engineering the Yeast Love Affair.Curr Biol. 2016 Jun 6;26(11):R463-6. doi: 10.1016/j.cub.2016.04.025. Curr Biol. 2016. PMID: 27269722
Similar articles
-
Cdc42 explores the cell periphery for mate selection in fission yeast.Curr Biol. 2013 Jan 7;23(1):42-7. doi: 10.1016/j.cub.2012.10.042. Epub 2012 Nov 29. Curr Biol. 2013. PMID: 23200991
-
Spatial focalization of pheromone/MAPK signaling triggers commitment to cell-cell fusion.Genes Dev. 2016 Oct 1;30(19):2226-2239. doi: 10.1101/gad.286922.116. Genes Dev. 2016. PMID: 27798845 Free PMC article.
-
Differential GAP requirement for Cdc42-GTP polarization during proliferation and sexual reproduction.J Cell Biol. 2018 Dec 3;217(12):4215-4229. doi: 10.1083/jcb.201806016. Epub 2018 Oct 2. J Cell Biol. 2018. PMID: 30279276 Free PMC article.
-
A focus on yeast mating: From pheromone signaling to cell-cell fusion.Semin Cell Dev Biol. 2023 Jan 15;133:83-95. doi: 10.1016/j.semcdb.2022.02.003. Epub 2022 Feb 9. Semin Cell Dev Biol. 2023. PMID: 35148940 Review.
-
The evolution of peptide mating pheromones in fission yeast.Curr Genet. 2019 Oct;65(5):1107-1111. doi: 10.1007/s00294-019-00968-w. Epub 2019 Apr 9. Curr Genet. 2019. PMID: 30968190 Review.
Cited by
-
Chemotactic movement of a polarity site enables yeast cells to find their mates.Proc Natl Acad Sci U S A. 2021 Jun 1;118(22):e2025445118. doi: 10.1073/pnas.2025445118. Proc Natl Acad Sci U S A. 2021. PMID: 34050026 Free PMC article.
-
Activation of Cdc42 GTPase upon CRY2-Induced Cortical Recruitment Is Antagonized by GAPs in Fission Yeast.Cells. 2020 Sep 12;9(9):2089. doi: 10.3390/cells9092089. Cells. 2020. PMID: 32932721 Free PMC article.
-
Unisexual reproduction promotes competition for mating partners in the global human fungal pathogen Cryptococcus deneoformans.PLoS Genet. 2019 Sep 19;15(9):e1008394. doi: 10.1371/journal.pgen.1008394. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31536509 Free PMC article.
-
The Multiple Functions of Rho GTPases in Fission Yeasts.Cells. 2021 Jun 7;10(6):1422. doi: 10.3390/cells10061422. Cells. 2021. PMID: 34200466 Free PMC article. Review.
-
Diverse mating phenotypes impact the spread of wtf meiotic drivers in Schizosaccharomyces pombe.Elife. 2021 Dec 13;10:e70812. doi: 10.7554/eLife.70812. Elife. 2021. PMID: 34895466 Free PMC article.
References
-
- Imai Y, Yamamoto M. The fission yeast mating pheromone P-factor: its molecular structure, gene structure, and ability to induce gene expression and G1 arrest in the mating partner. Genes Dev. 1994;8:328–338. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

