xGDBvm: A Web GUI-Driven Workflow for Annotating Eukaryotic Genomes in the Cloud
- PMID: 27020957
- PMCID: PMC4863385
- DOI: 10.1105/tpc.15.00933
xGDBvm: A Web GUI-Driven Workflow for Annotating Eukaryotic Genomes in the Cloud
Abstract
Genome-wide annotation of gene structure requires the integration of numerous computational steps. Currently, annotation is arguably best accomplished through collaboration of bioinformatics and domain experts, with broad community involvement. However, such a collaborative approach is not scalable at today's pace of sequence generation. To address this problem, we developed the xGDBvm software, which uses an intuitive graphical user interface to access a number of common genome analysis and gene structure tools, preconfigured in a self-contained virtual machine image. Once their virtual machine instance is deployed through iPlant's Atmosphere cloud services, users access the xGDBvm workflow via a unified Web interface to manage inputs, set program parameters, configure links to high-performance computing (HPC) resources, view and manage output, apply analysis and editing tools, or access contextual help. The xGDBvm workflow will mask the genome, compute spliced alignments from transcript and/or protein inputs (locally or on a remote HPC cluster), predict gene structures and gene structure quality, and display output in a public or private genome browser complete with accessory tools. Problematic gene predictions are flagged and can be reannotated using the integrated yrGATE annotation tool. xGDBvm can also be configured to append or replace existing data or load precomputed data. Multiple genomes can be annotated and displayed, and outputs can be archived for sharing or backup. xGDBvm can be adapted to a variety of use cases including de novo genome annotation, reannotation, comparison of different annotations, and training or teaching.
© 2016 American Society of Plant Biologists. All rights reserved.
Figures

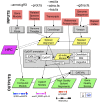



Similar articles
-
Laniakea: an open solution to provide Galaxy "on-demand" instances over heterogeneous cloud infrastructures.Gigascience. 2020 Apr 1;9(4):giaa033. doi: 10.1093/gigascience/giaa033. Gigascience. 2020. PMID: 32252069 Free PMC article.
-
PGen: large-scale genomic variations analysis workflow and browser in SoyKB.BMC Bioinformatics. 2016 Oct 6;17(Suppl 13):337. doi: 10.1186/s12859-016-1227-y. BMC Bioinformatics. 2016. PMID: 27766951 Free PMC article.
-
From the desktop to the grid: scalable bioinformatics via workflow conversion.BMC Bioinformatics. 2016 Mar 12;17:127. doi: 10.1186/s12859-016-0978-9. BMC Bioinformatics. 2016. PMID: 26968893 Free PMC article.
-
Improving data workflow systems with cloud services and use of open data for bioinformatics research.Brief Bioinform. 2018 Sep 28;19(5):1035-1050. doi: 10.1093/bib/bbx039. Brief Bioinform. 2018. PMID: 28419324 Free PMC article. Review.
-
Web tools for predictive toxicology model building.Expert Opin Drug Metab Toxicol. 2012 Jul;8(7):791-801. doi: 10.1517/17425255.2012.685158. Epub 2012 May 12. Expert Opin Drug Metab Toxicol. 2012. PMID: 22577953 Review.
Cited by
-
Sequence Assembly of Yarrowia lipolytica Strain W29/CLIB89 Shows Transposable Element Diversity.PLoS One. 2016 Sep 7;11(9):e0162363. doi: 10.1371/journal.pone.0162363. eCollection 2016. PLoS One. 2016. PMID: 27603307 Free PMC article.
References
-
- Abouelhoda M.I., Kurtz S., Ohlebusch E. (2002). The enhanced suffix array and its applications to genome analysis. In Second Workshop on Algorithms in Bioinformatics, R. Guigo and D. Gusfield, eds (Rome:Springer-Verlag; ), pp. 449–463.
-
- Dooley R., Vaughn M., Stanzione D., Terry S., Skidmore E. (2012). Software-as-a-Service: The iPlant Foundation API. In 5th IEEE Workshop on Many-Task Computing on Grids and Supercomputers (MTAGS) (IEEE; ).
-
- Foissac S., Gouzy J.P., Rombauts S., Mathé C., Amselem J., Sterck L., Van de Peer Y., Rouzé P., Schiex T. (2008). Genome annotation in plants and fungi: EuGene as a model platform. Curr. Bioinform. 3: 87–97.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

