Hepatitis C Virus Genotype 1 to 6 Protease Inhibitor Escape Variants: In Vitro Selection, Fitness, and Resistance Patterns in the Context of the Infectious Viral Life Cycle
- PMID: 27021330
- PMCID: PMC4879388
- DOI: 10.1128/AAC.02929-15
Hepatitis C Virus Genotype 1 to 6 Protease Inhibitor Escape Variants: In Vitro Selection, Fitness, and Resistance Patterns in the Context of the Infectious Viral Life Cycle
Abstract
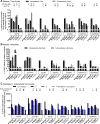
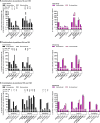
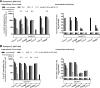
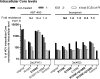
Hepatitis C virus (HCV) NS3 protease inhibitors (PIs) are important components of novel HCV therapy regimens. Studies of PI resistance initially focused on genotype 1. Therefore, knowledge about the determinants of PI resistance for the highly prevalent genotypes 2 to 6 remains limited. Using Huh7.5 cell culture-infectious HCV recombinants with genotype 1 to 6 NS3 protease, we identified protease positions 54, 155, and 156 as hot spots for the selection of resistance substitutions under treatment with the first licensed PIs, telaprevir and boceprevir. Treatment of a genotype 2 isolate with the newer PIs vaniprevir, faldaprevir, simeprevir, grazoprevir, paritaprevir, and deldeprevir identified positions 156 and 168 as hot spots for resistance; the Y56H substitution emerged for three newer PIs. Substitution selection also depended on the specific recombinant. The substitutions identified conferred cross-resistance to several PIs; however, most substitutions selected under telaprevir or boceprevir treatment conferred less resistance to certain newer PIs. In a single-cycle production assay, across genotypes, PI treatment primarily decreased viral replication, which was rescued by PI resistance substitutions. The substitutions identified resulted in differential effects on viral fitness, depending on the original recombinant and the substitution. Across genotypes, fitness impairment induced by resistance substitutions was due primarily to decreased replication. Most combinations of substitutions that were identified increased resistance or fitness. Combinations of resistance substitutions with fitness-compensating substitutions either rescued replication or compensated for decreased replication by increasing assembly. This comprehensive study provides insight into the selection patterns and effects of PI resistance substitutions for HCV genotypes 1 to 6 in the context of the infectious viral life cycle, which is of interest for clinical and virological HCV research.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Substitutions at NS3 Residue 155, 156, or 168 of Hepatitis C Virus Genotypes 2 to 6 Induce Complex Patterns of Protease Inhibitor Resistance.Antimicrob Agents Chemother. 2015 Dec;59(12):7426-36. doi: 10.1128/AAC.01953-15. Epub 2015 Sep 21. Antimicrob Agents Chemother. 2015. PMID: 26392503 Free PMC article.
-
HCV genotype 1-6 NS3 residue 80 substitutions impact protease inhibitor activity and promote viral escape.J Hepatol. 2019 Mar;70(3):388-397. doi: 10.1016/j.jhep.2018.10.031. Epub 2018 Nov 3. J Hepatol. 2019. PMID: 30395912
-
Implications of baseline polymorphisms for potential resistance to NS3 protease inhibitors in Hepatitis C virus genotypes 1a, 2b and 3a.Antiviral Res. 2013 Jul;99(1):12-7. doi: 10.1016/j.antiviral.2013.04.018. Epub 2013 May 3. Antiviral Res. 2013. PMID: 23648709
-
NS3 protease polymorphisms and genetic barrier to drug resistance of distinct hepatitis C virus genotypes from worldwide treatment-naïve subjects.J Viral Hepat. 2016 Nov;23(11):840-849. doi: 10.1111/jvh.12503. Epub 2016 Jan 18. J Viral Hepat. 2016. PMID: 26775769 Review.
-
Evolutionary Pathways to Persistence of Highly Fit and Resistant Hepatitis C Virus Protease Inhibitor Escape Variants.Hepatology. 2019 Sep;70(3):771-787. doi: 10.1002/hep.30647. Epub 2019 Jun 5. Hepatology. 2019. PMID: 30964552 Free PMC article. Review.
Cited by
-
Pre-existing resistance associated polymorphisms to NS3 protease inhibitors in treatment naïve HCV positive Pakistani patients.PLoS One. 2020 Apr 10;15(4):e0231480. doi: 10.1371/journal.pone.0231480. eCollection 2020. PLoS One. 2020. PMID: 32275694 Free PMC article.
-
Resistance detection and re-treatment options in hepatitis C virus-related chronic liver diseases after DAA-treatment failure.Infection. 2018 Dec;46(6):761-783. doi: 10.1007/s15010-018-1188-3. Epub 2018 Aug 6. Infection. 2018. PMID: 30084057 Review.
-
Virological Factors Associated with Failure to the Latest Generation of Direct Acting Agents (DAA) and Re-Treatment Strategy: A Narrative Review.Viruses. 2021 Mar 8;13(3):432. doi: 10.3390/v13030432. Viruses. 2021. PMID: 33800289 Free PMC article. Review.
-
Advancing high-throughput anti-HCV drug screening: a novel dual-reporter HCV replicon model with real-time monitoring.Res Pharm Sci. 2025 Feb 20;20(1):41-54. doi: 10.4103/RPS.RPS_249_23. eCollection 2025 Feb. Res Pharm Sci. 2025. PMID: 40190821 Free PMC article.
-
Metalloprotoporphyrin Inhibition of HCV NS3-4A Protease: Structure-Activity Relationships.Drug Des Devel Ther. 2020 Feb 24;14:757-771. doi: 10.2147/DDDT.S201089. eCollection 2020. Drug Des Devel Ther. 2020. PMID: 32158194 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

