Do Src Kinase and Caveolin Interact Directly with Na,K-ATPase?
- PMID: 27022017
- PMCID: PMC4882442
- DOI: 10.1074/jbc.M116.721084
Do Src Kinase and Caveolin Interact Directly with Na,K-ATPase?
Abstract
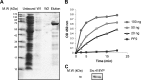
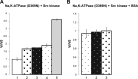
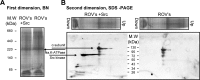
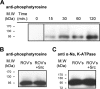
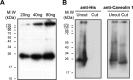
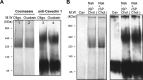
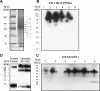
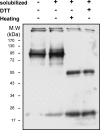
Much evidence points to a role of Na,K-ATPase in ouabain-dependent signal transduction. Based on experiments with different cell lines and native tissue membranes, a current hypothesis postulates direct interactions between the Na,K-ATPase and Src kinase (non-receptor tyrosine kinase). Na,K-ATPase is proposed to bind Src kinase and inhibit its activity, whereas ouabain, the specific Na,K-ATPase inhibitor, binds and stabilizes the E2 conformation, thus exposing the Src kinase domain and its active site Tyr-418 for activation. Ouabain-dependent signaling is thought to be mediated within caveolae by a complex consisting of Na,K-ATPase, caveolin, and Src kinase. In the current work, we have looked for direct interactions utilizing purified recombinant Na,K-ATPase (human α1β1FXYD1 or porcine α1D369Nβ1FXYD1) and purified human Src kinase and human caveolin 1 or interactions between these proteins in native membrane vesicles isolated from rabbit kidney. By several independent criteria and techniques, no stable interactions were detected between Na,K-ATPase and purified Src kinase. Na,K-ATPase was found to be a substrate for Src kinase phosphorylation at Tyr-144. Clear evidence for a direct interaction between purified human Na,K-ATPase and human caveolin was obtained, albeit with a low molar stoichiometry (1:15-30 caveolin 1/Na,K-ATPase). In native renal membranes, a specific caveolin 14-5 oligomer (95 kDa) was found to be in direct interaction with Na,K-ATPase. We inferred that a small fraction of the renal Na,K-ATPase molecules is in a ∼1:1 complex with a caveolin 14-5 oligomer. Thus, overall, whereas a direct caveolin 1/Na,K-ATPase interaction is confirmed, the lack of direct Src kinase/Na,K-ATPase binding requires reassessment of the mechanism of ouabain-dependent signaling.
Keywords: Na+/K+-ATPase; Src; caveolin; protein-protein interaction; signaling.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Kometiani P., Li J., Gnudi L., Kahn B. B., Askari A., and Xie Z. (1998) Multiple signal transduction pathways link Na+/K+-ATPase to growth-related genes in cardiac myocytes: the roles of Ras and mitogen-activated protein kinases. J. Biol. Chem. 273, 15249–15256 - PubMed
-
- Haas M., Wang H., Tian J., and Xie Z. (2002) Src-mediated inter-receptor cross-talk between the Na+/K+-ATPase and the epidermal growth factor receptor relays the signal from ouabain to mitogen-activated protein kinases. J. Biol. Chem. 277, 18694–18702 - PubMed
-
- Haas M., Askari A., and Xie Z. (2000) Involvement of Src and epidermal growth factor receptor in the signal-transducing function of Na+/K+-ATPase. J. Biol. Chem. 275, 27832–27837 - PubMed
-
- Aperia A. (2007) New roles for an old enzyme: Na,K-ATPase emerges as an interesting drug target. J. Intern. Med. 261, 44–52 - PubMed
-
- Schoner W. (2002) Endogenous cardiac glycosides, a new class of steroid hormones. Eur. J. Biochem. 269, 2440–2448 - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

