G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits
- PMID: 27022092
- PMCID: PMC4810302
- DOI: 10.1083/jcb.201508028
G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits
Erratum in
-
Correction: G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits.J Cell Biol. 2020 Jan 6;219(1):e20150802809202019c. doi: 10.1083/jcb.20150802809202019c. J Cell Biol. 2020. PMID: 31851327 Free PMC article. No abstract available.
Abstract
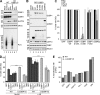
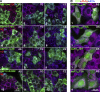
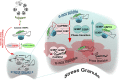
Mammalian stress granules (SGs) contain stalled translation preinitiation complexes that are assembled into discrete granules by specific RNA-binding proteins such as G3BP. We now show that cells lacking both G3BP1 and G3BP2 cannot form SGs in response to eukaryotic initiation factor 2α phosphorylation or eIF4A inhibition, but are still SG-competent when challenged with severe heat or osmotic stress. Rescue experiments using G3BP1 mutants show that phosphomimetic G3BP1-S149E fails to rescue SG formation, whereas G3BP1-F33W, a mutant unable to bind G3BP partner proteins Caprin1 or USP10, rescues SG formation. Caprin1/USP10 binding to G3BP is mutually exclusive: Caprin binding promotes, but USP10 binding inhibits, SG formation. G3BP interacts with 40S ribosomal subunits through its RGG motif, which is also required for G3BP-mediated SG formation. We propose that G3BP mediates the condensation of SGs by shifting between two different states that are controlled by the phosphorylation of S149 and by binding to Caprin1 or USP10.
© 2016 Kedersha et al.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

