The Roles of MDM2 and MDMX in Cancer
- PMID: 27022975
- PMCID: PMC6028239
- DOI: 10.1146/annurev-pathol-012414-040349
The Roles of MDM2 and MDMX in Cancer
Abstract
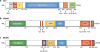
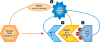
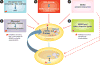
For more than 25 years, MDM2 and its homolog MDMX (also known as MDM4) have been shown to exert oncogenic activity. These two proteins are best understood as negative regulators of the p53 tumor suppressor, although they may have additional p53-independent roles. Understanding the dysregulation of MDM2 and MDMX in human cancers and how they function either together or separately in tumorigenesis may improve methods of diagnosis and for assessing prognosis. Targeting the proteins themselves, or their regulators, may be a promising therapeutic approach to treating some forms of cancer.
Keywords: MDM2; MDMX; cancer; p53; tumorigenesis.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

