Challenges in microbial ecology: building predictive understanding of community function and dynamics
- PMID: 27022995
- PMCID: PMC5113837
- DOI: 10.1038/ismej.2016.45
Challenges in microbial ecology: building predictive understanding of community function and dynamics
Abstract
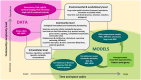
The importance of microbial communities (MCs) cannot be overstated. MCs underpin the biogeochemical cycles of the earth's soil, oceans and the atmosphere, and perform ecosystem functions that impact plants, animals and humans. Yet our ability to predict and manage the function of these highly complex, dynamically changing communities is limited. Building predictive models that link MC composition to function is a key emerging challenge in microbial ecology. Here, we argue that addressing this challenge requires close coordination of experimental data collection and method development with mathematical model building. We discuss specific examples where model-experiment integration has already resulted in important insights into MC function and structure. We also highlight key research questions that still demand better integration of experiments and models. We argue that such integration is needed to achieve significant progress in our understanding of MC dynamics and function, and we make specific practical suggestions as to how this could be achieved.
Conflict of interest statement
Dr Patrick B Warren holds equity (>$10k) in Unilever PLC. All other authors declare no conflict of interest.
Figures




References
-
- Agawin NSR, Rabouille S, Veldhuis MJW, Servatius L, Hol S, van Overzee HMJ et al. (2007). Competition and facilitation between unicellular nitrogen-fixing cyanobacteria and non-nitrogen-fixing phytoplankton species. Limnol Oceanogr 52: 2233–2248.
-
- Bai Y, Muller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528: 364–369. - PubMed
-
- Baranyi J, Tamplin ML. (2004). ComBase: a common database on microbial responses to food environments. J Food Prot 67: 1967–1971. - PubMed
Publication types
MeSH terms
Grants and funding
- BB/K003240/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/L015080/1/MRC_/Medical Research Council/United Kingdom
- MR/M50161X/1/MRC_/Medical Research Council/United Kingdom
- NC/K000683/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

