Species Identification of Bovine, Ovine and Porcine Type 1 Collagen; Comparing Peptide Mass Fingerprinting and LC-Based Proteomics Methods
- PMID: 27023524
- PMCID: PMC4848901
- DOI: 10.3390/ijms17040445
Species Identification of Bovine, Ovine and Porcine Type 1 Collagen; Comparing Peptide Mass Fingerprinting and LC-Based Proteomics Methods
Abstract
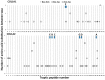
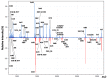
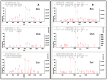
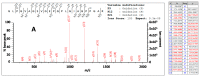
Collagen is one of the most ubiquitous proteins in the animal kingdom and the dominant protein in extracellular tissues such as bone, skin and other connective tissues in which it acts primarily as a supporting scaffold. It has been widely investigated scientifically, not only as a biomedical material for regenerative medicine, but also for its role as a food source for both humans and livestock. Due to the long-term stability of collagen, as well as its abundance in bone, it has been proposed as a source of biomarkers for species identification not only for heat- and pressure-rendered animal feed but also in ancient archaeological and palaeontological specimens, typically carried out by peptide mass fingerprinting (PMF) as well as in-depth liquid chromatography (LC)-based tandem mass spectrometric methods. Through the analysis of the three most common domesticates species, cow, sheep, and pig, this research investigates the advantages of each approach over the other, investigating sites of sequence variation with known functional properties of the collagen molecule. Results indicate that the previously identified species biomarkers through PMF analysis are not among the most variable type 1 collagen peptides present in these tissues, the latter of which can be detected by LC-based methods. However, it is clear that the highly repetitive sequence motif of collagen throughout the molecule, combined with the variability of the sites and relative abundance levels of hydroxylation, can result in high scoring false positive peptide matches using these LC-based methods. Additionally, the greater alpha 2(I) chain sequence variation, in comparison to the alpha 1(I) chain, did not appear to be specific to any particular functional properties, implying that intra-chain functional constraints on sequence variation are not as great as inter-chain constraints. However, although some of the most variable peptides were only observed in LC-based methods, until the range of publicly available collagen sequences improves, the simplicity of the PMF approach and suitable range of peptide sequence variation observed makes it the ideal method for initial taxonomic identification prior to further analysis by LC-based methods only when required.
Keywords: bone collagen; collagen function; hydroxylation; peptide mass fingerprinting; variability.
Figures



Similar articles
-
A Rapid and Simple LC-MS Method Using Collagen Marker Peptides for Identification of the Animal Source of Leather.J Agric Food Chem. 2016 Aug 3;64(30):6051-7. doi: 10.1021/acs.jafc.6b02132. Epub 2016 Jul 20. J Agric Food Chem. 2016. PMID: 27397145
-
Microwave-assisted acid hydrolysis of proteins combined with peptide fractionation and mass spectrometry analysis for characterizing protein terminal sequences.J Proteomics. 2014 Apr 4;100:68-78. doi: 10.1016/j.jprot.2013.10.014. Epub 2013 Oct 18. J Proteomics. 2014. PMID: 24145141
-
Microwave-assisted acid hydrolysis of proteins combined with liquid chromatography MALDI MS/MS for protein identification.J Am Soc Mass Spectrom. 2005 Apr;16(4):471-81. doi: 10.1016/j.jasms.2004.12.017. J Am Soc Mass Spectrom. 2005. PMID: 15792716
-
Applications of Tandem Mass Spectrometry (MS/MS) in Protein Analysis for Biomedical Research.Molecules. 2022 Apr 8;27(8):2411. doi: 10.3390/molecules27082411. Molecules. 2022. PMID: 35458608 Free PMC article. Review.
-
Protein sequence information by matrix-assisted laser desorption/ionization in-source decay mass spectrometry.Mass Spectrom Rev. 2007 Sep-Oct;26(5):672-82. doi: 10.1002/mas.20142. Mass Spectrom Rev. 2007. PMID: 17492750 Review.
Cited by
-
A Collagen Supplement Improves Skin Hydration, Elasticity, Roughness, and Density: Results of a Randomized, Placebo-Controlled, Blind Study.Nutrients. 2019 Oct 17;11(10):2494. doi: 10.3390/nu11102494. Nutrients. 2019. PMID: 31627309 Free PMC article. Clinical Trial.
-
Bioarchitectural Design of Bioactive Biopolymers: Structure-Function Paradigm for Diabetic Wound Healing.Biomimetics (Basel). 2024 May 4;9(5):275. doi: 10.3390/biomimetics9050275. Biomimetics (Basel). 2024. PMID: 38786486 Free PMC article. Review.
-
Cheminformatics-Based Design and Synthesis of Hydroxyapatite/Collagen Nanocomposites for Biomedical Applications.Polymers (Basel). 2023 Dec 27;16(1):85. doi: 10.3390/polym16010085. Polymers (Basel). 2023. PMID: 38201750 Free PMC article.
-
Proteomics as a tool to gain next level insights into photo-crosslinkable biopolymer modifications.Bioact Mater. 2022 Jan 23;17:204-220. doi: 10.1016/j.bioactmat.2022.01.023. eCollection 2022 Nov. Bioact Mater. 2022. PMID: 35386456 Free PMC article.
-
In-silico molecular analysis and blocking of the viral G protein of Nipah virus interacting with ephrin B2 and B3 receptor by using peptide mass fingerprinting.Front Bioinform. 2025 Jun 25;5:1526566. doi: 10.3389/fbinf.2025.1526566. eCollection 2025. Front Bioinform. 2025. PMID: 40635997 Free PMC article.
References
-
- Whitmore R., Jones H., Windus W., Naghski J. Preparation of hide collagen for food. J. Am. Leather Chem. Assoc. 1970;65:382–389.
-
- Gómez-Guillén M., Giménez B., López-Caballero M., Montero M. Functional and bioactive properties of collagen and gelatin from alternative sources: A review. Food Hydrocoll. 2011;25:1813–1827. doi: 10.1016/j.foodhyd.2011.02.007. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

