Fecal Microbial Transplants Reduce Antibiotic-resistant Genes in Patients With Recurrent Clostridium difficile Infection
- PMID: 27025836
- PMCID: PMC4885654
- DOI: 10.1093/cid/ciw185
Fecal Microbial Transplants Reduce Antibiotic-resistant Genes in Patients With Recurrent Clostridium difficile Infection
Abstract
Background: Recurrent Clostridium difficile infection (RCDI) is associated with repeated antibiotic treatment and the enhanced growth of antibiotic-resistant microbes. This study tested the hypothesis that patients with RCDI would harbor large numbers of antibiotic-resistant microbes and that fecal microbiota transplantation (FMT) would reduce the number of antibiotic-resistant genes.
Methods: In a single center study, patients with RCDI (n = 20) received FMT from universal donors via colonoscopy. Stool samples were collected from donors (n = 3) and patients prior to and following FMT. DNA was extracted and shotgun metagenomics performed. Results as well as assembled libraries from a healthy cohort (n = 87) obtained from the Human Microbiome Project were aligned against the NCBI bacterial taxonomy database and the Comprehensive Antibiotic Resistance Database. Results were corroborated through a DNA microarray containing 354 antibiotic resistance (ABR) genes.
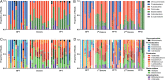
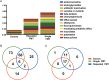
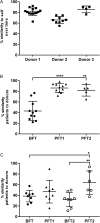
Results: RCDI patients had a greater number and diversity of ABR genes compared with donors and healthy controls. Beta-lactam, multidrug efflux pumps, fluoroquinolone, and antibiotic inactivation ABR genes were increased in RCDI patients, although donors primarily had tetracycline resistance. RCDI patients were dominated by Proteobacteria with Escherichia coli and Klebsiella most prevalent. FMT resulted in a resolution of symptoms that correlated directly with a decreased number and diversity of ABR genes and increased Bacteroidetes and Firmicutes with reduced Proteobacteria. ABR gene profiles were maintained in recipients for up to a year following FMT.
Conclusions: RCDI patients have increased numbers of antibiotic-resistant organisms. FMT is effective in the eradication of pathogenic antibiotic-resistant organisms and elimination of ABR genes.
Keywords: C. difficile; antibiotic resistance; antibiotics; colitis; intestinal microbiome.
© The Author 2016. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures


Comment in
-
Editorial Commentary: The Dawning of Microbiome Remediation for Addressing Antibiotic Resistance.Clin Infect Dis. 2016 Jun 15;62(12):1487-8. doi: 10.1093/cid/ciw187. Epub 2016 Mar 29. Clin Infect Dis. 2016. PMID: 27025827 Free PMC article. No abstract available.
-
Reduction of Antibiotic Resistance Genes in Intestinal Microbiota of Patients With Recurrent Clostridium difficile Infection After Fecal Microbiota Transplantation.Clin Infect Dis. 2016 Sep 1;63(5):710-1. doi: 10.1093/cid/ciw390. Epub 2016 Jun 17. Clin Infect Dis. 2016. PMID: 27317794 No abstract available.
-
Reply to Jouhten et al.Clin Infect Dis. 2016 Sep 1;63(5):711-2. doi: 10.1093/cid/ciw393. Epub 2016 Jun 17. Clin Infect Dis. 2016. PMID: 27317795 No abstract available.
References
-
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature 2000; 405:299–304. - PubMed
-
- Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 2010; 8:251–9. - PubMed
-
- McFarland LV. Antibiotic-associated diarrhea: epidemiology, trends and treatment. Future Microbiol 2008; 3:563–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

