A Perfect Storm: Impact of Genomic Variation and Serial Vaccination on Low Influenza Vaccine Effectiveness During the 2014-2015 Season
- PMID: 27025838
- PMCID: PMC4901864
- DOI: 10.1093/cid/ciw176
A Perfect Storm: Impact of Genomic Variation and Serial Vaccination on Low Influenza Vaccine Effectiveness During the 2014-2015 Season
Abstract
Background: The 2014-2015 influenza season was distinguished by an epidemic of antigenically-drifted A(H3N2) viruses and vaccine components identical to 2013-2014. We report 2014-2015 vaccine effectiveness (VE) from Canada and explore contributing agent-host factors.
Methods: VE against laboratory-confirmed influenza was derived using a test-negative design among outpatients with influenza-like illness. Sequencing identified amino acid mutations at key antigenic sites of the viral hemagglutinin protein.
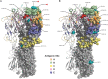
Results: Overall, 815/1930 (42%) patients tested influenza-positive: 590 (72%) influenza A and 226 (28%) influenza B. Most influenza A viruses with known subtype were A(H3N2) (570/577; 99%); 409/460 (89%) sequenced viruses belonged to genetic clade 3C.2a and 39/460 (8%) to clade 3C.3b. Dominant clade 3C.2a viruses bore the pivotal mutations F159Y (a cluster-transition position) and K160T (a predicted gain of glycosylation) compared to the mismatched clade 3C.1 vaccine. VE against A(H3N2) was -17% (95% confidence interval [CI], -50% to 9%) overall with clade-specific VE of -13% (95% CI, -51% to 15%) for clade 3C.2a but 52% (95% CI, -17% to 80%) for clade 3C.3b. VE against A(H3N2) was 53% (95% CI, 10% to 75%) for patients vaccinated in 2014-2015 only, significantly lower at -32% (95% CI, -75% to 0%) if also vaccinated in 2013-2014 and -54% (95% CI, -108% to -14%) if vaccinated each year since 2012-2013. VE against clade-mismatched B(Yamagata) viruses was 42% (95% CI, 10% to 62%) with less-pronounced reduction from prior vaccination compared to A(H3N2).
Conclusions: Variation in the viral genome and negative effects of serial vaccination likely contributed to poor influenza vaccine performance in 2014-2015.
Keywords: antigenic drift; genomics; influenza vaccines; sentinel surveillance; vaccine effectiveness.
© The Author 2016. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures


References
-
- Centers for Disease Control and Prevention. Influenza (flu). Past weekly surveillance reports. Available at: http://www.cdc.gov/flu/weekly/pastreports.htm Accessed 4 April 2016.
-
- Public Health Agency of Canada. FluWatch. Weekly reports 2014–2015 season. Available at: http://www.phac-aspc.gc.ca/fluwatch/14-15/index-eng.php Accessed 4 April 2016.
-
- World Health Organization. Recommended composition of influenza virus vaccines for use in the 2014–2015 northern hemisphere influenza season. Geneva: WHO, 2014. Available at: http://www.who.int/influenza/vaccines/virus/recommendations/2014_15_nort... Accessed 4 April 2016.
-
- World Health Organization. Recommended composition of influenza virus vaccines for use in the 2015–2016 northern hemisphere influenza season. Geneva: WHO, 2015. Available at: http://www.who.int/influenza/vaccines/virus/recommendations/2015_16_nort... Accessed 4 April 2016.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

