PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability
- PMID: 27026615
- PMCID: PMC5070514
- DOI: 10.1093/jamia/ocv202
PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability
Abstract
Objective: Health care generated data have become an important source for clinical and genomic research. Often, investigators create and iteratively refine phenotype algorithms to achieve high positive predictive values (PPVs) or sensitivity, thereby identifying valid cases and controls. These algorithms achieve the greatest utility when validated and shared by multiple health care systems.Materials and Methods We report the current status and impact of the Phenotype KnowledgeBase (PheKB, http://phekb.org), an online environment supporting the workflow of building, sharing, and validating electronic phenotype algorithms. We analyze the most frequent components used in algorithms and their performance at authoring institutions and secondary implementation sites.
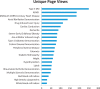
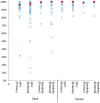
Results: As of June 2015, PheKB contained 30 finalized phenotype algorithms and 62 algorithms in development spanning a range of traits and diseases. Phenotypes have had over 3500 unique views in a 6-month period and have been reused by other institutions. International Classification of Disease codes were the most frequently used component, followed by medications and natural language processing. Among algorithms with published performance data, the median PPV was nearly identical when evaluated at the authoring institutions (n = 44; case 96.0%, control 100%) compared to implementation sites (n = 40; case 97.5%, control 100%).
Discussion: These results demonstrate that a broad range of algorithms to mine electronic health record data from different health systems can be developed with high PPV, and algorithms developed at one site are generally transportable to others.
Conclusion: By providing a central repository, PheKB enables improved development, transportability, and validity of algorithms for research-grade phenotypes using health care generated data.
Keywords: clinical research; electronic health records; electronic phenotyping; genomic research; natural language processing.
© The Author 2016. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

