JAM: A Scalable Bayesian Framework for Joint Analysis of Marginal SNP Effects
- PMID: 27027514
- PMCID: PMC4817278
- DOI: 10.1002/gepi.21953
JAM: A Scalable Bayesian Framework for Joint Analysis of Marginal SNP Effects
Abstract
Recently, large scale genome-wide association study (GWAS) meta-analyses have boosted the number of known signals for some traits into the tens and hundreds. Typically, however, variants are only analysed one-at-a-time. This complicates the ability of fine-mapping to identify a small set of SNPs for further functional follow-up. We describe a new and scalable algorithm, joint analysis of marginal summary statistics (JAM), for the re-analysis of published marginal summary statistics under joint multi-SNP models. The correlation is accounted for according to estimates from a reference dataset, and models and SNPs that best explain the complete joint pattern of marginal effects are highlighted via an integrated Bayesian penalized regression framework. We provide both enumerated and Reversible Jump MCMC implementations of JAM and present some comparisons of performance. In a series of realistic simulation studies, JAM demonstrated identical performance to various alternatives designed for single region settings. In multi-region settings, where the only multivariate alternative involves stepwise selection, JAM offered greater power and specificity. We also present an application to real published results from MAGIC (meta-analysis of glucose and insulin related traits consortium) - a GWAS meta-analysis of more than 15,000 people. We re-analysed several genomic regions that produced multiple significant signals with glucose levels 2 hr after oral stimulation. Through joint multivariate modelling, JAM was able to formally rule out many SNPs, and for one gene, ADCY5, suggests that an additional SNP, which transpired to be more biologically plausible, should be followed up with equal priority to the reported index.
Keywords: GWAS meta-analysis; fine-mapping; glucose; insulin; variable selection.
© 2016 The Authors Genetic Epidemiology Published by Wiley Periodicals, Inc.
Figures
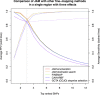


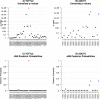

References
-
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265. - PubMed
-
- Bottolo L, Richardson S. 2010. Evolutionary stochastic search for Bayesian model exploration. Bayesian Anal 5(3):583–618.
Publication types
MeSH terms
Substances
Grants and funding
- G1001799/MRC_/Medical Research Council/United Kingdom
- R01 ES016813/ES/NIEHS NIH HHS/United States
- R01 CA140561/CA/NCI NIH HHS/United States
- MR/N01104X/1/MRC_/Medical Research Council/United Kingdom
- R01 CA132839/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01CA140561/CA/NCI NIH HHS/United States
- U01 HG004726/HG/NHGRI NIH HHS/United States
- 076113/WT_/Wellcome Trust/United Kingdom
- MC_UP_0801/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00002/10/MRC_/Medical Research Council/United Kingdom
- 076113/C/04/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

