The Identification of Novel Diagnostic Marker Genes for the Detection of Beer Spoiling Pediococcus damnosus Strains Using the BlAst Diagnostic Gene findEr
- PMID: 27028007
- PMCID: PMC4814128
- DOI: 10.1371/journal.pone.0152747
The Identification of Novel Diagnostic Marker Genes for the Detection of Beer Spoiling Pediococcus damnosus Strains Using the BlAst Diagnostic Gene findEr
Abstract
As the number of bacterial genomes increases dramatically, the demand for easy to use tools with transparent functionality and comprehensible output for applied comparative genomics grows as well. We present BlAst Diagnostic Gene findEr (BADGE), a tool for the rapid prediction of diagnostic marker genes (DMGs) for the differentiation of bacterial groups (e.g. pathogenic / nonpathogenic). DMG identification settings can be modified easily and installing and running BADGE does not require specific bioinformatics skills. During the BADGE run the user is informed step by step about the DMG finding process, thus making it easy to evaluate the impact of chosen settings and options. On the basis of an example with relevance for beer brewing, being one of the oldest biotechnological processes known, we show a straightforward procedure, from phenotyping, genome sequencing, assembly and annotation, up to a discriminant marker gene PCR assay, making comparative genomics a means to an end. The value and the functionality of BADGE were thoroughly examined, resulting in the successful identification and validation of an outstanding novel DMG (fabZ) for the discrimination of harmless and harmful contaminations of Pediococcus damnosus, which can be applied for spoilage risk determination in breweries. Concomitantly, we present and compare five complete P. damnosus genomes sequenced in this study, finding that the ability to produce the unwanted, spoilage associated off-flavor diacetyl is a plasmid encoded trait in this important beer spoiling species.
Conflict of interest statement
Figures
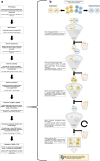
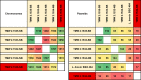
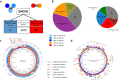
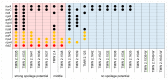
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

