Detecting Gene Rearrangements in Patient Populations Through a 2-Step Diagnostic Test Comprised of Rapid IHC Enrichment Followed by Sensitive Next-Generation Sequencing
- PMID: 27028240
- PMCID: PMC5553231
- DOI: 10.1097/PAI.0000000000000360
Detecting Gene Rearrangements in Patient Populations Through a 2-Step Diagnostic Test Comprised of Rapid IHC Enrichment Followed by Sensitive Next-Generation Sequencing
Abstract
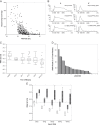
Targeted therapy combined with companion diagnostics has led to the advancement of next-generation sequencing (NGS) for detection of molecular alterations. However, using a diagnostic test to identify patient populations with low prevalence molecular alterations, such as gene rearrangements, poses efficiency, and cost challenges. To address this, we have developed a 2-step diagnostic test to identify NTRK1, NTRK2, NTRK3, ROS1, and ALK rearrangements in formalin-fixed paraffin-embedded clinical specimens. This test is comprised of immunohistochemistry screening using a pan-receptor tyrosine kinase cocktail of antibodies to identify samples expressing TrkA (encoded by NTRK1), TrkB (encoded by NTRK2), TrkC (encoded by NTRK3), ROS1, and ALK followed by an RNA-based anchored multiplex polymerase chain reaction NGS assay. We demonstrate that the NGS assay is accurate and reproducible in identification of gene rearrangements. Furthermore, implementation of an RNA quality control metric to assess the presence of amplifiable nucleic acid input material enables a measure of confidence when an NGS result is negative for gene rearrangements. Finally, we demonstrate that performing a pan-receptor tyrosine kinase immunohistochemistry staining enriches detection of the patient population for gene rearrangements from 4% to 9% and has a 100% negative predictive value. Together, this 2-step assay is an efficient method for detection of gene rearrangements in both clinical testing and studies of archival formalin-fixed paraffin-embedded specimens.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

