Genetic diversity analysis of two commercial breeds of pigs using genomic and pedigree data
- PMID: 27029213
- PMCID: PMC4812646
- DOI: 10.1186/s12711-016-0203-3
Genetic diversity analysis of two commercial breeds of pigs using genomic and pedigree data
Abstract
Background: Genetic improvement in livestock populations can be achieved without significantly affecting genetic diversity if mating systems and selection decisions take genetic relationships among individuals into consideration. The objective of this study was to examine the genetic diversity of two commercial breeds of pigs. Genotypes from 1168 Landrace (LA) and 1094 Large White (LW) animals from a commercial breeding program in Brazil were obtained using the Illumina PorcineSNP60 Beadchip. Inbreeding estimates based on pedigree (F x) and genomic information using runs of homozygosity (F ROH) and the single nucleotide polymorphisms (SNP) by SNP inbreeding coefficient (F SNP) were obtained. Linkage disequilibrium (LD), correlation of linkage phase (r) and effective population size (N e ) were also estimated.
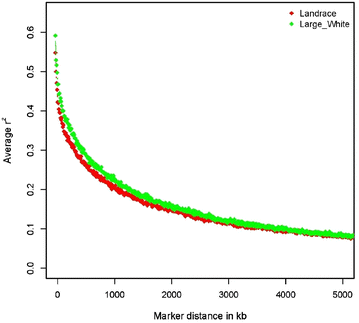
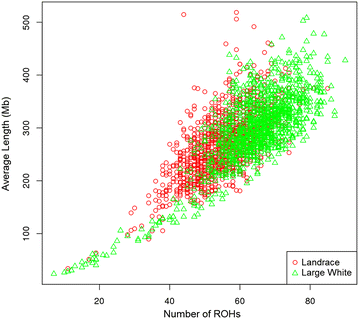
Results: Estimates of inbreeding obtained with pedigree information were lower than those obtained with genomic data in both breeds. We observed that the extent of LD was slightly larger at shorter distances between SNPs in the LW population than in the LA population, which indicates that the LW population was derived from a smaller N e . Estimates of N e based on genomic data were equal to 53 and 40 for the current populations of LA and LW, respectively. The correlation of linkage phase between the two breeds was equal to 0.77 at distances up to 50 kb, which suggests that genome-wide association and selection should be performed within breed. Although selection intensities have been stronger in the LA breed than in the LW breed, levels of genomic and pedigree inbreeding were lower for the LA than for the LW breed.
Conclusions: The use of genomic data to evaluate population diversity in livestock animals can provide new and more precise insights about the effects of intense selection for production traits. Resulting information and knowledge can be used to effectively increase response to selection by appropriately managing the rate of inbreeding, minimizing negative effects of inbreeding depression and therefore maintaining desirable levels of genetic diversity.
Figures



Similar articles
-
Genome-wide linkage disequilibrium and genetic diversity in five populations of Australian domestic sheep.Genet Sel Evol. 2015 Nov 24;47:90. doi: 10.1186/s12711-015-0169-6. Genet Sel Evol. 2015. PMID: 26602211 Free PMC article.
-
Characterization and management of long runs of homozygosity in parental nucleus lines and their associated crossbred progeny.Genet Sel Evol. 2016 Nov 24;48(1):91. doi: 10.1186/s12711-016-0269-y. Genet Sel Evol. 2016. PMID: 27884108 Free PMC article.
-
Comparative evaluation of genomic inbreeding parameters in seven commercial and autochthonous pig breeds.Animal. 2020 May;14(5):910-920. doi: 10.1017/S175173111900332X. Epub 2020 Jan 13. Animal. 2020. PMID: 31928538
-
Runs of homozygosity: current knowledge and applications in livestock.Anim Genet. 2017 Jun;48(3):255-271. doi: 10.1111/age.12526. Epub 2016 Dec 1. Anim Genet. 2017. PMID: 27910110 Review.
-
Invited review: Inbreeding in the genomics era: Inbreeding, inbreeding depression, and management of genomic variability.J Dairy Sci. 2017 Aug;100(8):6009-6024. doi: 10.3168/jds.2017-12787. Epub 2017 Jun 7. J Dairy Sci. 2017. PMID: 28601448 Review.
Cited by
-
Evolutionary pressures rendered by animal husbandry practices for avian influenza viruses to adapt to humans.iScience. 2022 Mar 1;25(4):104005. doi: 10.1016/j.isci.2022.104005. eCollection 2022 Apr 15. iScience. 2022. PMID: 35313691 Free PMC article. Review.
-
Genomic structure of a crossbred Landrace pig population.PLoS One. 2019 Feb 28;14(2):e0212266. doi: 10.1371/journal.pone.0212266. eCollection 2019. PLoS One. 2019. PMID: 30818344 Free PMC article.
-
The coefficients of inbreeding revealed by ROH study among inbred individuals belonging to each type of the first cousin marriage: A preliminary report from North India.Genes Genomics. 2023 Jun;45(6):813-825. doi: 10.1007/s13258-023-01367-9. Epub 2023 Feb 20. Genes Genomics. 2023. PMID: 36807878
-
Identification of Genetic Regions Associated with Scrotal Hernias in a Commercial Swine Herd.Vet Sci. 2018 Jan 27;5(1):15. doi: 10.3390/vetsci5010015. Vet Sci. 2018. PMID: 29382056 Free PMC article.
-
Whole-genome sequencing reveals patterns of runs of homozygosity underlying genetic diversity and selection in domestic rabbits.BMC Genomics. 2025 Apr 29;26(1):425. doi: 10.1186/s12864-025-11616-8. BMC Genomics. 2025. PMID: 40301718 Free PMC article.
References
-
- ABCS 2013. http://www.abcs.org.br/images/pdf/registro.pdf. Accessed 14 Oct 2014.
-
- Weigel KA. Controlling inbreeding in modern breeding programs. J Dairy Sci. 2001;84:E177–E184. doi: 10.3168/jds.S0022-0302(01)70213-5. - DOI
-
- Burrow HM. The effects of inbreeding in beef cattle. Anim Breed Abstr. 1993;61:737–751.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

