Molecular Mechanisms and Clinical Impact of Acquired and Intrinsic Fosfomycin Resistance
- PMID: 27029300
- PMCID: PMC4790336
- DOI: 10.3390/antibiotics2020217
Molecular Mechanisms and Clinical Impact of Acquired and Intrinsic Fosfomycin Resistance
Abstract
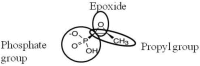
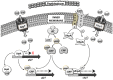
Bacterial infections caused by antibiotic-resistant isolates have become a major health problem in recent years, since they are very difficult to treat, leading to an increase in morbidity and mortality. Fosfomycin is a broad-spectrum bactericidal antibiotic that inhibits cell wall biosynthesis in both Gram-negative and Gram-positive bacteria. This antibiotic has a unique mechanism of action and inhibits the initial step in peptidoglycan biosynthesis by blocking the enzyme, MurA. Fosfomycin has been used successfully for the treatment of urinary tract infections for a long time, but the increased emergence of antibiotic resistance has made fosfomycin a suitable candidate for the treatment of infections caused by multidrug-resistant pathogens, especially in combination with other therapeutic partners. The acquisition of fosfomycin resistance could threaten the reintroduction of this antibiotic for the treatment of bacterial infection. Here, we analyse the mechanism of action and molecular mechanisms for the development of fosfomycin resistance, including the modification of the antibiotic target, reduced antibiotic uptake and antibiotic inactivation. In addition, we describe the role of each pathway in clinical isolates.
Keywords: fosfomycin resistance; molecular mechanisms.
Figures


References
-
- Falagas M.E., Grammatikos A.P., Michalopoulos A. Potential of old-generation antibiotics to address current need for new antibiotics. Expert Rev. Anti. Infect. Ther. 2008;6:593–600. - PubMed
-
- Hendlin D., Stapley E.O., Jackson M., Wallick H., Miller A.K., Wolf F.J., Miller T.W., Chaiet L., Kahan F.M., Foltz E.L., et al. Phosphonomycin, a new antibiotic produced by strains of Streptomyces. Science. 1969;166:122–123. - PubMed
-
- Kahan F.M., Kahan J.S., Cassidy P.J., Kropp H. The mechanism of action of fosfomycin (phosphonomycin) Ann. NY Acad. Sci. 1974;235:364–386. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

