pH Induced Conformational Transitions in the Transforming Growth Factor β-Induced Protein (TGFβIp) Associated Corneal Dystrophy Mutants
- PMID: 27030015
- PMCID: PMC4814907
- DOI: 10.1038/srep23836
pH Induced Conformational Transitions in the Transforming Growth Factor β-Induced Protein (TGFβIp) Associated Corneal Dystrophy Mutants
Abstract
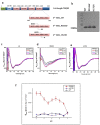
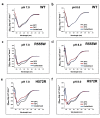
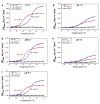
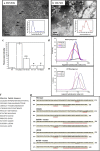
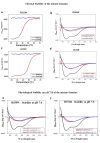
Most stromal corneal dystrophies are associated with aggregation and deposition of the mutated transforming growth factor-β induced protein (TGFβIp). The 4(th)_FAS1 domain of TGFβIp harbors ~80% of the mutations that forms amyloidogenic and non-amyloidogenic aggregates. To understand the mechanism of aggregation and the differences between the amyloidogenic and non-amyloidogenic phenotypes, we expressed the 4(th)_FAS1 domains of TGFβIp carrying the mutations R555W (non-amyloidogenic) and H572R (amyloidogenic) along with the wild-type (WT). R555W was more susceptible to acidic pH compared to H572R and displayed varying chemical stabilities with decreasing pH. Thermal denaturation studies at acidic pH showed that while WT did not undergo any conformational transition, the mutants exhibited a clear pH-dependent irreversible conversion from αβ conformation to β-sheet oligomers. The β-oligomers of both mutants were stable at physiological temperature and pH. Electron microscopy and dynamic light scattering studies showed that β-oligomers of H572R were larger compared to R555W. The β-oligomers of both mutants were cytotoxic to primary human corneal stromal fibroblast (pHCSF) cells. The β-oligomers of both mutants exhibit variations in their morphologies, sizes, thermal and chemical stabilities, aggregation patterns and cytotoxicities.
Figures



Similar articles
-
Expression, purification and characterization of fourth FAS1 domain of TGFβIp-associated corneal dystrophic mutants.Protein Expr Purif. 2012 Jul;84(1):108-15. doi: 10.1016/j.pep.2012.04.018. Epub 2012 May 2. Protein Expr Purif. 2012. PMID: 22575305
-
Structural and Functional Implications of Human Transforming Growth Factor β-Induced Protein, TGFBIp, in Corneal Dystrophies.Structure. 2017 Nov 7;25(11):1740-1750.e2. doi: 10.1016/j.str.2017.09.001. Epub 2017 Oct 5. Structure. 2017. PMID: 28988748
-
Mutation-Induced Deamidation of Corneal Dystrophy-Related Transforming Growth Factor β-Induced Protein.Biochemistry. 2017 Dec 12;56(49):6470-6480. doi: 10.1021/acs.biochem.7b00668. Epub 2017 Nov 22. Biochemistry. 2017. PMID: 29140698
-
Clinical and genetic aspects of the TGFBI-associated corneal dystrophies.Ocul Surf. 2014 Oct;12(4):234-51. doi: 10.1016/j.jtos.2013.12.002. Epub 2014 Jul 18. Ocul Surf. 2014. PMID: 25284770 Review.
-
Biochemical properties and aggregation propensity of transforming growth factor-induced protein (TGFBIp) and the amyloid forming mutants.Ocul Surf. 2015 Jan;13(1):9-25. doi: 10.1016/j.jtos.2014.04.003. Epub 2014 Oct 8. Ocul Surf. 2015. PMID: 25557343 Review.
Cited by
-
Case of progressive hyperopia due to flattening of cornea.Am J Ophthalmol Case Rep. 2018 Feb 24;10:169-171. doi: 10.1016/j.ajoc.2018.02.013. eCollection 2018 Jun. Am J Ophthalmol Case Rep. 2018. PMID: 29780931 Free PMC article.
-
Effect of position-specific single-point mutations and biophysical characterization of amyloidogenic peptide fragments identified from lattice corneal dystrophy patients.Biochem J. 2017 May 9;474(10):1705-1725. doi: 10.1042/BCJ20170125. Biochem J. 2017. PMID: 28381645 Free PMC article.
-
Fascinating Fasciclins: A Surprisingly Widespread Family of Proteins that Mediate Interactions between the Cell Exterior and the Cell Surface.Int J Mol Sci. 2018 May 31;19(6):1628. doi: 10.3390/ijms19061628. Int J Mol Sci. 2018. PMID: 29857505 Free PMC article. Review.
-
Effect of osmolytes on in-vitro aggregation properties of peptides derived from TGFBIp.Sci Rep. 2020 Mar 4;10(1):4011. doi: 10.1038/s41598-020-60944-0. Sci Rep. 2020. PMID: 32132634 Free PMC article.
-
Alternatives for Uranyl Acetate Negative Staining With Respect to the Resolution Obtained After Single Particle Analysis of Erythrocruorin.Microsc Res Tech. 2025 Sep;88(9):2381-2391. doi: 10.1002/jemt.24865. Epub 2025 Mar 31. Microsc Res Tech. 2025. PMID: 40165377 Free PMC article.
References
-
- Surguchev A. & Surguchov A. Conformational diseases: looking into the eyes. Brain Res. Bull 81, 12–24 (2010). - PubMed
-
- Lakshminarayanan R. et al. Biochemical properties and aggregation propensity of transforming growth factor-induced protein (TGFβIp) and the amyloid forming mutants. Ocul Surf. 13(1), 9–25 (2015). - PubMed
-
- Kitahama S. et al. Expression of fibrillins and other microfibril-associated proteins in human bone and osteoblast-like cells. Bone 27, 61–67 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

