Roles of Us8A and Its Phosphorylation Mediated by Us3 in Herpes Simplex Virus 1 Pathogenesis
- PMID: 27030266
- PMCID: PMC4886770
- DOI: 10.1128/JVI.00446-16
Roles of Us8A and Its Phosphorylation Mediated by Us3 in Herpes Simplex Virus 1 Pathogenesis
Abstract
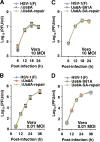
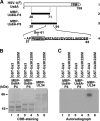
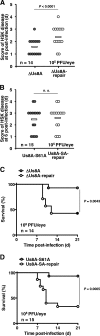
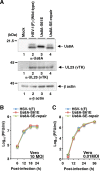
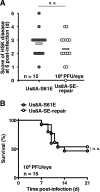
The herpes simplex virus 1 (HSV-1) Us8A gene overlaps the gene that encodes glycoprotein E (gE). Previous studies have investigated the roles of Us8A in HSV-1 infection using null mutations in Us8A and gE; therefore, the role of Us8A remains to be elucidated. In this study, we investigated the function of Us8A and its phosphorylation at serine 61 (Ser-61), which we recently identified as a phosphorylation site by mass spectrometry-based phosphoproteomic analysis of HSV-1-infected cells, in HSV-1 pathogenesis. We observed that (i) the phosphorylation of Us8A Ser-61 in infected cells was dependent on the activity of the virus-encoded Us3 protein kinase; (ii) the Us8A null mutant virus exhibited a 10-fold increase in the 50% lethal dose for virulence in the central nervous system (CNS) of mice following intracranial infection compared with a repaired virus; (iii) replacement of Ser-61 with alanine (S61A) in Us8A had little effect on virulence in the CNS of mice following intracranial infection, whereas it significantly reduced the mortality of mice following ocular infection to levels similar to the Us8A null mutant virus; (iv) the Us8A S61A mutation also significantly reduced viral yields in mice following ocular infection, mainly in the trigeminal ganglia and brains; and (v) a phosphomimetic mutation at Us8A Ser-61 restored wild-type viral yields and virulence. Collectively, these results indicate that Us8A is a novel HSV-1 virulence factor and suggest that the Us3-mediated phosphorylation of Us8A Ser-61 regulates Us8A function for viral invasion into the CNS from peripheral sites.
Importance: The DNA genomes of viruses within the subfamily Alphaherpesvirinae are divided into unique long (UL) and unique short (Us) regions. Us regions contain alphaherpesvirus-specific genes. Recently, high-throughput sequencing of ocular isolates of HSV-1 showed that Us8A was the most highly conserved of 13 herpes simplex virus 1 (HSV-1) genes mapped to the Us region, suggesting Us8A may have an important role in the HSV-1 life cycle. However, the specific role of Us8A in HSV-1 infection remains to be elucidated. Here, we show that Us8A is a virulence factor for HSV-1 infection in mice, and the function of Us8A for viral invasion into the central nervous system from peripheral sites is regulated by Us3-mediated phosphorylation of the protein at Ser-61. This is the first study to report the significance of Us8A and its regulation in HSV-1 infection.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Phosphorylation of a herpes simplex virus 1 dUTPase by a viral protein kinase, Us3, dictates viral pathogenicity in the central nervous system but not at the periphery.J Virol. 2014 Mar;88(5):2775-85. doi: 10.1128/JVI.03300-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352467 Free PMC article.
-
Impact of the changes in substrate specificity of herpes simplex virus 1 protein kinase Us3 on viral infection in vitro and in vivo.J Virol. 2025 Jul 22;99(7):e0040025. doi: 10.1128/jvi.00400-25. Epub 2025 Jun 30. J Virol. 2025. PMID: 40586577 Free PMC article.
-
Role of herpes simplex virus 1 Us3 in viral neuroinvasiveness.Microbiol Immunol. 2014 Jan;58(1):31-7. doi: 10.1111/1348-0421.12108. Microbiol Immunol. 2014. PMID: 24200420
-
Us3 Protein Kinase Encoded by HSV: The Precise Function and Mechanism on Viral Life Cycle.Adv Exp Med Biol. 2018;1045:45-62. doi: 10.1007/978-981-10-7230-7_3. Adv Exp Med Biol. 2018. PMID: 29896662 Review.
-
[Molecular mechanism by which Us3 protein kinase regulates the pathogenicity of herpes simplex virus type-1].Uirusu. 2016;66(1):83-90. doi: 10.2222/jsv.66.83. Uirusu. 2016. PMID: 28484184 Review. Japanese.
Cited by
-
The Us2 Gene Product of Herpes Simplex Virus 2 modulates NF-κB activation by targeting TAK1.Sci Rep. 2017 Aug 21;7(1):8396. doi: 10.1038/s41598-017-08856-4. Sci Rep. 2017. PMID: 28827540 Free PMC article.
-
Impact of the interaction between herpes simplex virus 1 ICP22 and FACT on viral gene expression and pathogenesis.J Virol. 2024 Aug 20;98(8):e0073724. doi: 10.1128/jvi.00737-24. Epub 2024 Jul 17. J Virol. 2024. PMID: 39016551 Free PMC article.
-
Genotypic and Phenotypic Diversity of Herpes Simplex Virus 2 within the Infected Neonatal Population.mSphere. 2019 Feb 27;4(1):e00590-18. doi: 10.1128/mSphere.00590-18. mSphere. 2019. PMID: 30814317 Free PMC article.
-
Roles of the Phosphorylation of Herpes Simplex Virus 1 UL51 at a Specific Site in Viral Replication and Pathogenicity.J Virol. 2018 Aug 29;92(18):e01035-18. doi: 10.1128/JVI.01035-18. Print 2018 Sep 15. J Virol. 2018. PMID: 29976672 Free PMC article.
-
Impaired endocytosis and accumulation in early endosomal compartments defines herpes simplex virus-mediated disruption of the nonclassical MHC class I-related molecule MR1.J Biol Chem. 2024 Oct;300(10):107748. doi: 10.1016/j.jbc.2024.107748. Epub 2024 Sep 12. J Biol Chem. 2024. PMID: 39260697 Free PMC article.
References
-
- Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses, p 1823–1897. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott-Williams &Wilkins, Philadelphia, PA.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

