The high mobility group protein HMO1 functions as a linker histone in yeast
- PMID: 27030801
- PMCID: PMC4812653
- DOI: 10.1186/s13072-016-0062-8
The high mobility group protein HMO1 functions as a linker histone in yeast
Abstract
Background: Eukaryotic chromatin consists of nucleosome core particles connected by linker DNA of variable length. Histone H1 associates with the linker DNA to stabilize the higher-order chromatin structure and to modulate the ability of regulatory factors to access their nucleosomal targets. In Saccharomyces cerevisiae, the protein with greatest sequence similarity to H1 is Hho1p. However, during vegetative growth, hho1∆ cells do not show any discernible cell growth defects or the changes in bulk chromatin structure that are characteristic of chromatin from multicellular eukaryotes in which H1 is depleted. In contrast, the yeast high mobility group (HMGB) protein HMO1 has been reported to compact chromatin, as evidenced by increased nuclease sensitivity in hmo1∆ cells. HMO1 has an unusual domain architecture compared to vertebrate HMGB proteins in that the HMG domains are followed by a lysine-rich extension instead of an acidic domain. We address here the hypothesis that HMO1 serves the role of H1 in terms of chromatin compaction and that this function requires the lysine-rich extension.
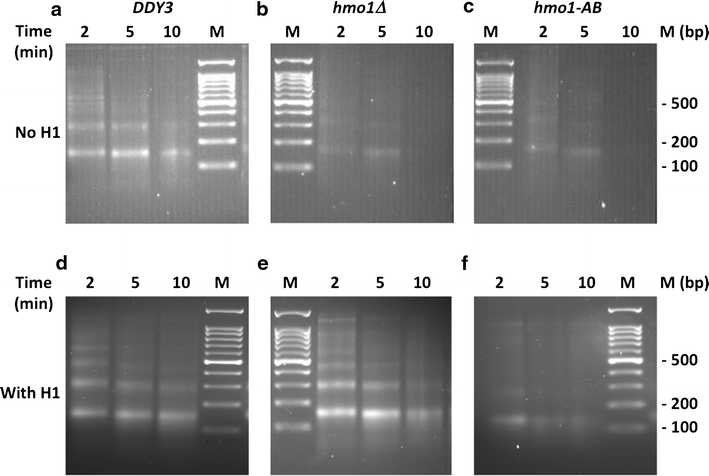
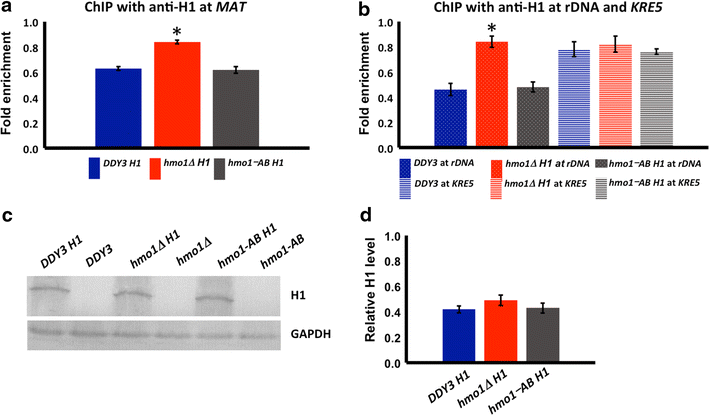
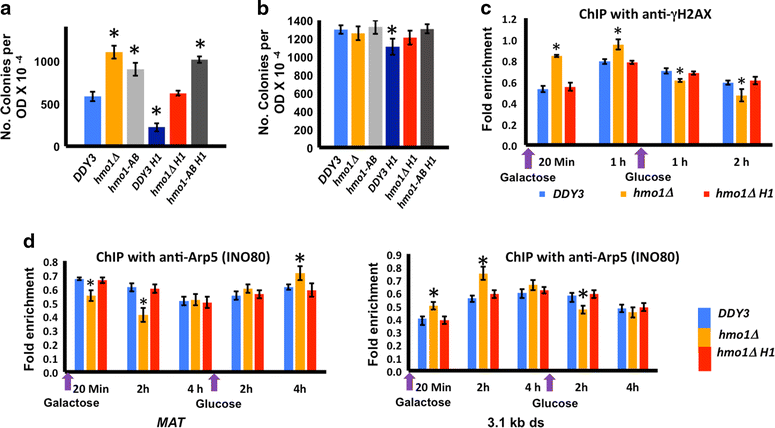
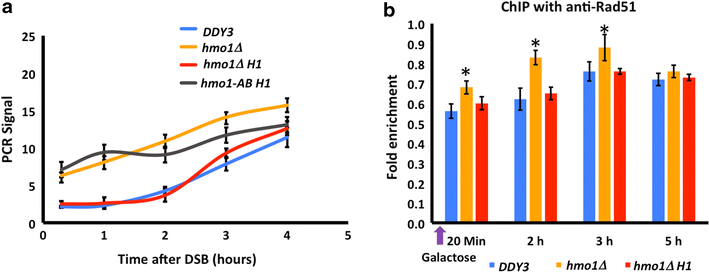
Results: We show here that HMO1 fulfills this function of a linker histone. For histone H1, chromatin compaction requires its basic C-terminal domain, and we find that the same pertains to HMO1, as deletion of its C-terminal lysine-rich extension renders chromatin nuclease sensitive. On rDNA, deletion of both HMO1 and Hho1p is required for significantly increased nuclease sensitivity. Expression of human histone H1 completely reverses the nuclease sensitivity characteristic of chromatin isolated from hmo1∆ cells. While chromatin remodeling events associated with repair of DNA double-strand breaks occur faster in the more dynamic chromatin environment created by the hmo1 deletion, expression of human histone H1 results in chromatin remodeling and double-strand break repair similar to that observed in wild-type cells.
Conclusion: Our data suggest that S. cerevisiae HMO1 protects linker DNA from nuclease digestion, a property also characteristic of mammalian linker histone H1. Notably, association with HMO1 creates a less dynamic chromatin environment that depends on its lysine-rich domain. That HMO1 has linker histone function has implications for investigations of chromatin structure and function as well as for evolution of proteins with roles in chromatin compaction.
Keywords: Chromatin; Chromatin immunoprecipitation; Double-strand break repair; Hho1p; High mobility group protein; Histone H1.
Figures





Similar articles
-
Yeast HMO1: Linker Histone Reinvented.Microbiol Mol Biol Rev. 2016 Nov 30;81(1):e00037-16. doi: 10.1128/MMBR.00037-16. Print 2017 Mar. Microbiol Mol Biol Rev. 2016. PMID: 27903656 Free PMC article. Review.
-
Single-molecule study reveals Hmo1, not Hho1, promotes chromatin assembly in budding yeast.mBio. 2023 Aug 31;14(4):e0099323. doi: 10.1128/mbio.00993-23. Epub 2023 Jul 11. mBio. 2023. PMID: 37432033 Free PMC article.
-
Yeast high mobility group protein HMO1 stabilizes chromatin and is evicted during repair of DNA double strand breaks.Nucleic Acids Res. 2015 Jul 13;43(12):5759-70. doi: 10.1093/nar/gkv498. Epub 2015 May 15. Nucleic Acids Res. 2015. PMID: 25979266 Free PMC article.
-
Hmo1: A versatile member of the high mobility group box family of chromosomal architecture proteins.World J Biol Chem. 2024 Aug 12;15(1):97938. doi: 10.4331/wjbc.v15.i1.97938. World J Biol Chem. 2024. PMID: 39156122 Free PMC article. Review.
-
The linker histone Hho1 modulates the activity of ATP-dependent chromatin remodeling complexes.Biochim Biophys Acta Gene Regul Mech. 2022 Jan;1865(1):194781. doi: 10.1016/j.bbagrm.2021.194781. Epub 2021 Dec 25. Biochim Biophys Acta Gene Regul Mech. 2022. PMID: 34963628
Cited by
-
Inhibition of transcription leads to rewiring of locus-specific chromatin proteomes.Genome Res. 2020 Apr;30(4):635-646. doi: 10.1101/gr.256255.119. Epub 2020 Mar 18. Genome Res. 2020. PMID: 32188699 Free PMC article.
-
Specialization of the chromatin remodeler RSC to mobilize partially-unwrapped nucleosomes.Elife. 2020 Jun 4;9:e58130. doi: 10.7554/eLife.58130. Elife. 2020. PMID: 32496195 Free PMC article.
-
Control of RNA polymerase II-transcribed genes by direct binding of TOR kinase.Curr Genet. 2018 Feb;64(1):131-135. doi: 10.1007/s00294-017-0738-z. Epub 2017 Aug 22. Curr Genet. 2018. PMID: 28831551 Review.
-
Resetting the Yeast Epigenome with Human Nucleosomes.Cell. 2017 Dec 14;171(7):1508-1519.e13. doi: 10.1016/j.cell.2017.10.043. Epub 2017 Nov 30. Cell. 2017. PMID: 29198523 Free PMC article.
-
Yeast HMO1: Linker Histone Reinvented.Microbiol Mol Biol Rev. 2016 Nov 30;81(1):e00037-16. doi: 10.1128/MMBR.00037-16. Print 2017 Mar. Microbiol Mol Biol Rev. 2016. PMID: 27903656 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

