Investigating the Proton Donor in the NO Reductase from Paracoccus denitrificans
- PMID: 27030968
- PMCID: PMC4816578
- DOI: 10.1371/journal.pone.0152745
Investigating the Proton Donor in the NO Reductase from Paracoccus denitrificans
Erratum in
-
Correction: Investigating the Proton Donor in the NO Reductase from Paracoccus denitrificans.PLoS One. 2016 May 20;11(5):e0155365. doi: 10.1371/journal.pone.0155365. eCollection 2016. PLoS One. 2016. PMID: 27203242 Free PMC article.
Abstract
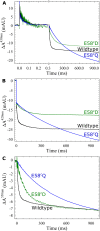
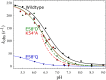
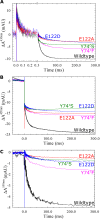
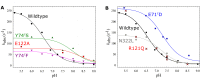
Variant nomenclature: the variants were made in the NorB subunit if not indicated by the superscript c, which are variants in the NorC subunit (e.g. E122A = exchange of Glu-122 in NorB for an Ala, E71cD; exchange of Glu-71 in NorC for an Asp). Bacterial NO reductases (NORs) are integral membrane proteins from the heme-copper oxidase superfamily. Most heme-copper oxidases are proton-pumping enzymes that reduce O2 as the last step in the respiratory chain. With electrons from cytochrome c, NO reductase (cNOR) from Paracoccus (P.) denitrificans reduces NO to N2O via the following reaction: 2NO+2e-+2H+→N2O+H2O. Although this reaction is as exergonic as O2-reduction, cNOR does not contribute to the electrochemical gradient over the membrane. This means that cNOR does not pump protons and that the protons needed for the reaction are taken from the periplasmic side of the membrane (since the electrons are donated from this side). We previously showed that the P. denitrificans cNOR uses a single defined proton pathway with residues Glu-58 and Lys-54 from the NorC subunit at the entrance. Here we further strengthened the evidence in support of this pathway. Our further aim was to define the continuation of the pathway and the immediate proton donor for the active site. To this end, we investigated the region around the calcium-binding site and both propionates of heme b3 by site directed mutagenesis. Changing single amino acids in these areas often had severe effects on cNOR function, with many variants having a perturbed active site, making detailed analysis of proton transfer properties difficult. Our data does however indicate that the calcium ligation sphere and the region around the heme b3 propionates are important for proton transfer and presumably contain the proton donor. The possible evolutionary link between the area for the immediate donor in cNOR and the proton loading site (PLS) for pumped protons in oxygen-reducing heme-copper oxidases is discussed.
Conflict of interest statement
Figures




Similar articles
-
The nitric-oxide reductase from Paracoccus denitrificans uses a single specific proton pathway.J Biol Chem. 2013 Oct 18;288(42):30626-30635. doi: 10.1074/jbc.M113.497347. Epub 2013 Sep 6. J Biol Chem. 2013. PMID: 24014024 Free PMC article.
-
A pathway for protons in nitric oxide reductase from Paracoccus denitrificans.Biochim Biophys Acta. 2007 May;1767(5):362-73. doi: 10.1016/j.bbabio.2007.03.006. Epub 2007 Mar 16. Biochim Biophys Acta. 2007. PMID: 17466934
-
Comparative genomics and site-directed mutagenesis support the existence of only one input channel for protons in the C-family (cbb3 oxidase) of heme-copper oxygen reductases.Biochemistry. 2007 Sep 4;46(35):9963-72. doi: 10.1021/bi700659y. Epub 2007 Aug 4. Biochemistry. 2007. PMID: 17676874
-
Proton transfer in bacterial nitric oxide reductase.Biochem Soc Trans. 2006 Feb;34(Pt 1):188-90. doi: 10.1042/BST0340188. Biochem Soc Trans. 2006. PMID: 16417518 Review.
-
Cytochrome c oxidase (heme aa3) from Paracoccus denitrificans: analysis of mutations in putative proton channels of subunit I.J Bioenerg Biomembr. 1998 Feb;30(1):89-97. doi: 10.1023/a:1020515713103. J Bioenerg Biomembr. 1998. PMID: 9623810 Review.
Cited by
-
Oxygen Activation and Energy Conservation by Cytochrome c Oxidase.Chem Rev. 2018 Mar 14;118(5):2469-2490. doi: 10.1021/acs.chemrev.7b00664. Epub 2018 Jan 19. Chem Rev. 2018. PMID: 29350917 Free PMC article. Review.
-
Proton Pumping and Non-Pumping Terminal Respiratory Oxidases: Active Sites Intermediates of These Molecular Machines and Their Derivatives.Int J Mol Sci. 2021 Oct 7;22(19):10852. doi: 10.3390/ijms221910852. Int J Mol Sci. 2021. PMID: 34639193 Free PMC article. Review.
-
Correction: Investigating the Proton Donor in the NO Reductase from Paracoccus denitrificans.PLoS One. 2016 May 20;11(5):e0155365. doi: 10.1371/journal.pone.0155365. eCollection 2016. PLoS One. 2016. PMID: 27203242 Free PMC article.
-
Characterization of the quinol-dependent nitric oxide reductase from the pathogen Neisseria meningitidis, an electrogenic enzyme.Sci Rep. 2018 Feb 26;8(1):3637. doi: 10.1038/s41598-018-21804-0. Sci Rep. 2018. PMID: 29483528 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

