Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing
- PMID: 27031497
- PMCID: PMC4895203
- DOI: 10.1038/nprot.2016.043
Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing
Abstract
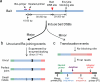
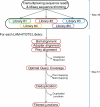
Unbiased, high-throughput assays for detecting and quantifying DNA double-stranded breaks (DSBs) across the genome in mammalian cells will facilitate basic studies of the mechanisms that generate and repair endogenous DSBs. They will also enable more applied studies, such as those to evaluate the on- and off-target activities of engineered nucleases. Here we describe a linear amplification-mediated high-throughput genome-wide sequencing (LAM-HTGTS) method for the detection of genome-wide 'prey' DSBs via their translocation in cultured mammalian cells to a fixed 'bait' DSB. Bait-prey junctions are cloned directly from isolated genomic DNA using LAM-PCR and unidirectionally ligated to bridge adapters; subsequent PCR steps amplify the single-stranded DNA junction library in preparation for Illumina Miseq paired-end sequencing. A custom bioinformatics pipeline identifies prey sequences that contribute to junctions and maps them across the genome. LAM-HTGTS differs from related approaches because it detects a wide range of broken end structures with nucleotide-level resolution. Familiarity with nucleic acid methods and next-generation sequencing analysis is necessary for library generation and data interpretation. LAM-HTGTS assays are sensitive, reproducible, relatively inexpensive, scalable and straightforward to implement with a turnaround time of <1 week.
Figures





Similar articles
-
Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases.Nat Biotechnol. 2015 Feb;33(2):179-86. doi: 10.1038/nbt.3101. Epub 2014 Dec 15. Nat Biotechnol. 2015. PMID: 25503383 Free PMC article.
-
END-seq: An Unbiased, High-Resolution, and Genome-Wide Approach to Map DNA Double-Strand Breaks and Resection in Human Cells.Methods Mol Biol. 2021;2153:9-31. doi: 10.1007/978-1-0716-0644-5_2. Methods Mol Biol. 2021. PMID: 32840769
-
Improved HTGTS for CRISPR/Cas9 off-target detection.Bio Protoc. 2019 May 5;9(9):e3229. doi: 10.21769/BioProtoc.3229. eCollection 2019 May 5. Bio Protoc. 2019. PMID: 33655015 Free PMC article.
-
Ionizing radiation and genetic risks. XVII. Formation mechanisms underlying naturally occurring DNA deletions in the human genome and their potential relevance for bridging the gap between induced DNA double-strand breaks and deletions in irradiated germ cells.Mutat Res. 2013 Oct-Dec;753(2):114-130. doi: 10.1016/j.mrrev.2013.07.003. Epub 2013 Aug 12. Mutat Res. 2013. PMID: 23948232 Review.
-
Defining genome-wide CRISPR-Cas genome-editing nuclease activity with GUIDE-seq.Nat Protoc. 2021 Dec;16(12):5592-5615. doi: 10.1038/s41596-021-00626-x. Epub 2021 Nov 12. Nat Protoc. 2021. PMID: 34773119 Free PMC article. Review.
Cited by
-
CRISPR/Cas9-induced structural variations expand in T lymphocytes in vivo.Nucleic Acids Res. 2022 Oct 28;50(19):11128-11137. doi: 10.1093/nar/gkac887. Nucleic Acids Res. 2022. PMID: 36243978 Free PMC article.
-
Expanding the editable genome and CRISPR-Cas9 versatility using DNA cutting-free gene targeting based on in trans paired nicking.Nucleic Acids Res. 2020 Jan 24;48(2):974-995. doi: 10.1093/nar/gkz1121. Nucleic Acids Res. 2020. PMID: 31799604 Free PMC article.
-
Comparative analysis of CRISPR off-target discovery tools following ex vivo editing of CD34+ hematopoietic stem and progenitor cells.Mol Ther. 2023 Apr 5;31(4):1074-1087. doi: 10.1016/j.ymthe.2023.02.011. Epub 2023 Feb 15. Mol Ther. 2023. PMID: 36793210 Free PMC article.
-
Switch Tandem Repeats Influence the Choice of the Alternative End-Joining Pathway in Immunoglobulin Class Switch Recombination.Front Immunol. 2022 May 16;13:870933. doi: 10.3389/fimmu.2022.870933. eCollection 2022. Front Immunol. 2022. PMID: 35651614 Free PMC article.
-
Induction of recurrent break cluster genes in neural progenitor cells differentiated from embryonic stem cells in culture.Proc Natl Acad Sci U S A. 2020 May 12;117(19):10541-10546. doi: 10.1073/pnas.1922299117. Epub 2020 Apr 24. Proc Natl Acad Sci U S A. 2020. PMID: 32332169 Free PMC article.
References
-
- Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv. Immunol. 2012;116:1–49. - PubMed
-
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

