DTW-MIC Coexpression Networks from Time-Course Data
- PMID: 27031641
- PMCID: PMC4816347
- DOI: 10.1371/journal.pone.0152648
DTW-MIC Coexpression Networks from Time-Course Data
Abstract
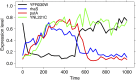
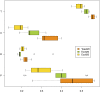
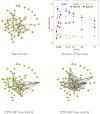
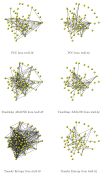
When modeling coexpression networks from high-throughput time course data, Pearson Correlation Coefficient (PCC) is one of the most effective and popular similarity functions. However, its reliability is limited since it cannot capture non-linear interactions and time shifts. Here we propose to overcome these two issues by employing a novel similarity function, Dynamic Time Warping Maximal Information Coefficient (DTW-MIC), combining a measure taking care of functional interactions of signals (MIC) and a measure identifying time lag (DTW). By using the Hamming-Ipsen-Mikhailov (HIM) metric to quantify network differences, the effectiveness of the DTW-MIC approach is demonstrated on a set of four synthetic and one transcriptomic datasets, also in comparison to TimeDelay ARACNE and Transfer Entropy.
Conflict of interest statement
Figures

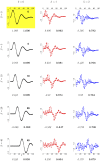
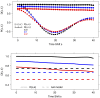
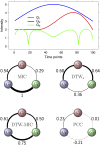
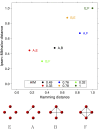

References
-
- De Smet R, Marchal K. Advantages and limitations of current network inference methods. Nat Rev Microbiol. 2010;8(10):717–729. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

