Thermodynamic Basis of Selectivity in the Interactions of Tissue Inhibitors of Metalloproteinases N-domains with Matrix Metalloproteinases-1, -3, and -14
- PMID: 27033700
- PMCID: PMC4900279
- DOI: 10.1074/jbc.M116.720250
Thermodynamic Basis of Selectivity in the Interactions of Tissue Inhibitors of Metalloproteinases N-domains with Matrix Metalloproteinases-1, -3, and -14
Abstract
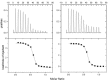
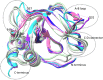
The four tissue inhibitors of metalloproteinases (TIMPs) are potent inhibitors of the many matrixins (MMPs), except that TIMP1 weakly inhibits some MMPs, including MMP14. The broad-spectrum inhibition of MMPs by TIMPs and their N-domains (NTIMPs) is consistent with the previous isothermal titration calorimetric finding that their interactions are entropy-driven but differ in contributions from solvent and conformational entropy (ΔSsolv, ΔSconf), estimated using heat capacity changes (ΔCp). Selective engineered NTIMPs have potential applications for treating MMP-related diseases, including cancer and cardiomyopathy. Here we report isothermal titration calorimetric studies of the effects of selectivity-modifying mutations in NTIMP1 and NTIMP2 on the thermodynamics of their interactions with MMP1, MMP3, and MMP14. The weak inhibition of MMP14 by NTIMP1 reflects a large conformational entropy penalty for binding. The T98L mutation, peripheral to the NTIMP1 reactive site, enhances binding by increasing ΔSsolv but also reduces ΔSconf However, the same mutation increases NTIMP1 binding to MMP3 in an interaction that has an unusual positive ΔCp This indicates a decrease in solvent entropy compensated by increased conformational entropy, possibly reflecting interactions involving alternative conformers. The NTIMP2 mutant, S2D/S4A is a selective MMP1 inhibitor through electrostatic effects of a unique MMP-1 arginine. Asp-2 increases reactive site polarity, reducing ΔCp, but increases conformational entropy to maintain strong binding to MMP1. There is a strong negative correlation between ΔSsolv and ΔSconf for all characterized interactions, but the data for each MMP have characteristic ranges, reflecting intrinsic differences in the structures and dynamics of their free and inhibitor-bound forms.
Keywords: calorimetry; conformational change; enzyme inhibitor; matrix metalloproteinase (MMP); protein dynamic; protein engineering; protein-protein interaction; thermodynamics; tissue inhibitor of metalloproteinase (TIMP); zinc.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
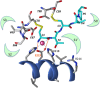
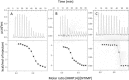
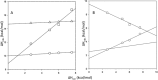
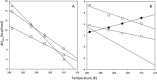

References
-
- Velázquez Campoy A., and Freire E. (2005) ITC in the post-genomic era….? Priceless. Biophys. Chem. 115, 115–124 - PubMed
-
- Murphy K. P., Freire E. (1992) Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv. Protein Chem. 43, 313–361 - PubMed
-
- Spolar R. S., Livingstone J. R., and Record M. T. Jr. (1992) Use of liquid hydrocarbon and amide transfer data to estimate contributions to thermodynamic functions of protein folding from the removal of nonpolar and polar surface from water. Biochemistry 31, 3947–3955 - PubMed
-
- Baker B. M., and Murphy K. P. (1998) Prediction of binding energetics from structure using empirical parameterization. Methods Enzymol. 295, 294–315 - PubMed
-
- Arumugam S., Gao G., Patton B. L., Semenchenko V., Brew K., and Van Doren S. R. (2003) Increased backbone mobility in β-barrel enhances entropy gain driving binding of N-TIMP-1 to MMP-3. J. Mol. Biol. 327, 719–734 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

