Genome-wide analysis of Musashi-2 targets reveals novel functions in governing epithelial cell migration
- PMID: 27034466
- PMCID: PMC4857000
- DOI: 10.1093/nar/gkw207
Genome-wide analysis of Musashi-2 targets reveals novel functions in governing epithelial cell migration
Abstract
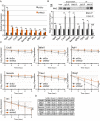
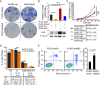
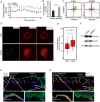
The Musashi-2 (Msi2) RNA-binding protein maintains stem cell self-renewal and promotes oncogenesis by enhancing cell proliferation in hematopoietic and gastrointestinal tissues. However, it is unclear how Msi2 recognizes and regulates mRNA targets in vivo and whether Msi2 primarily controls cell growth in all cell types. Here we identified Msi2 targets with HITS-CLIP and revealed that Msi2 primarily recognizes mRNA 3'UTRs at sites enriched in multiple copies of UAG motifs in epithelial progenitor cells. RNA-seq and ribosome profiling demonstrated that Msi2 promotes targeted mRNA decay without affecting translation efficiency. Unexpectedly, the most prominent Msi2 targets identified are key regulators that govern cell motility with a high enrichment in focal adhesion and extracellular matrix-receptor interaction, in addition to regulators of cell growth and survival. Loss of Msi2 stimulates epithelial cell migration, increases the number of focal adhesions and also compromises cell growth. These findings provide new insights into the molecular mechanisms of Msi2's recognition and repression of targets and uncover a key function of Msi2 in restricting epithelial cell migration.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Nakamura M., Okano H., Blendy J.A., Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

