In Silico Analysis of Arabidopsis thaliana Peroxisomal 6-Phosphogluconate Dehydrogenase
- PMID: 27034898
- PMCID: PMC4789532
- DOI: 10.1155/2016/3482760
In Silico Analysis of Arabidopsis thaliana Peroxisomal 6-Phosphogluconate Dehydrogenase
Abstract
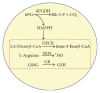
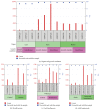
NADPH, whose regeneration is critical for reductive biosynthesis and detoxification pathways, is an essential component in cell redox homeostasis. Peroxisomes are subcellular organelles with a complex biochemical machinery involved in signaling and stress processes by molecules such as hydrogen peroxide (H2O2) and nitric oxide (NO). NADPH is required by several peroxisomal enzymes involved in β-oxidation, NO, and glutathione (GSH) generation. Plants have various NADPH-generating dehydrogenases, one of which is 6-phosphogluconate dehydrogenase (6PGDH). Arabidopsis contains three 6PGDH genes that probably are encoded for cytosolic, chloroplastic/mitochondrial, and peroxisomal isozymes, although their specific functions remain largely unknown. This study focuses on the in silico analysis of the biochemical characteristics and gene expression of peroxisomal 6PGDH (p6PGDH) with the aim of understanding its potential function in the peroxisomal NADPH-recycling system. The data show that a group of plant 6PGDHs contains an archetypal type 1 peroxisomal targeting signal (PTS), while in silico gene expression analysis using affymetrix microarray data suggests that Arabidopsis p6PGDH appears to be mainly involved in xenobiotic response, growth, and developmental processes.
Figures
Similar articles
-
Peroxisomal NADP-isocitrate dehydrogenase is required for Arabidopsis stomatal movement.Protoplasma. 2016 Mar;253(2):403-15. doi: 10.1007/s00709-015-0819-0. Epub 2015 Apr 19. Protoplasma. 2016. PMID: 25894616
-
Peroxisomal plant metabolism - an update on nitric oxide, Ca2+ and the NADPH recycling network.J Cell Sci. 2018 Jan 29;131(2):jcs202978. doi: 10.1242/jcs.202978. J Cell Sci. 2018. PMID: 28775155 Review.
-
Glyphosate-induced oxidative stress in Arabidopsis thaliana affecting peroxisomal metabolism and triggers activity in the oxidative phase of the pentose phosphate pathway (OxPPP) involved in NADPH generation.J Plant Physiol. 2017 Nov;218:196-205. doi: 10.1016/j.jplph.2017.08.007. Epub 2017 Aug 31. J Plant Physiol. 2017. PMID: 28888161
-
Nitric oxide and hydrogen sulfide modulate the NADPH-generating enzymatic system in higher plants.J Exp Bot. 2021 Feb 11;72(3):830-847. doi: 10.1093/jxb/eraa440. J Exp Bot. 2021. PMID: 32945878 Review.
-
A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes.Biochem J. 1998 Mar 1;330 ( Pt 2)(Pt 2):777-84. doi: 10.1042/bj3300777. Biochem J. 1998. PMID: 9480890 Free PMC article.
Cited by
-
The Arabidopsis Plastidial Glucose-6-Phosphate Transporter GPT1 is Dually Targeted to Peroxisomes via the Endoplasmic Reticulum.Plant Cell. 2020 May;32(5):1703-1726. doi: 10.1105/tpc.19.00959. Epub 2020 Feb 28. Plant Cell. 2020. PMID: 32111666 Free PMC article.
-
Peroxisomal Proteome Mining of Sweet Pepper (Capsicum annuum L.) Fruit Ripening Through Whole Isobaric Tags for Relative and Absolute Quantitation Analysis.Front Plant Sci. 2022 May 9;13:893376. doi: 10.3389/fpls.2022.893376. eCollection 2022. Front Plant Sci. 2022. PMID: 35615143 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of the Thioredoxin (Trx) Gene Family Reveals Its Role in Leaf Rust Resistance in Wheat (Triticum aestivum L.).Front Genet. 2022 Mar 25;13:836030. doi: 10.3389/fgene.2022.836030. eCollection 2022. Front Genet. 2022. PMID: 35401694 Free PMC article.
-
A bioinformatics approach to characterize a hypothetical protein Q6S8D9_SARS of SARS-CoV.Genomics Inform. 2023 Mar;21(1):e3. doi: 10.5808/gi.22021. Epub 2023 Mar 31. Genomics Inform. 2023. PMID: 37037461 Free PMC article.
-
Evidence for dual targeting control of Arabidopsis 6-phosphogluconate dehydrogenase isoforms by N-terminal phosphorylation.J Exp Bot. 2024 May 20;75(10):2848-2866. doi: 10.1093/jxb/erae077. J Exp Bot. 2024. PMID: 38412416 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

