Excessive Osteocytic Fgf23 Secretion Contributes to Pyrophosphate Accumulation and Mineralization Defect in Hyp Mice
- PMID: 27035636
- PMCID: PMC4818020
- DOI: 10.1371/journal.pbio.1002427
Excessive Osteocytic Fgf23 Secretion Contributes to Pyrophosphate Accumulation and Mineralization Defect in Hyp Mice
Abstract
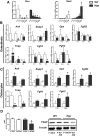
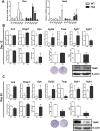
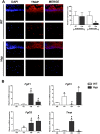
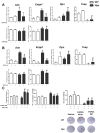
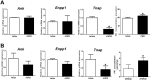
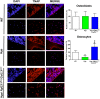
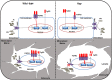
X-linked hypophosphatemia (XLH) is the most frequent form of inherited rickets in humans caused by mutations in the phosphate-regulating gene with homologies to endopeptidases on the X-chromosome (PHEX). Hyp mice, a murine homologue of XLH, are characterized by hypophosphatemia, inappropriately low serum vitamin D levels, increased serum fibroblast growth factor-23 (Fgf23), and osteomalacia. Although Fgf23 is known to be responsible for hypophosphatemia and reduced vitamin D hormone levels in Hyp mice, its putative role as an auto-/paracrine osteomalacia-causing factor has not been explored. We recently reported that Fgf23 is a suppressor of tissue nonspecific alkaline phosphatase (Tnap) transcription via FGF receptor-3 (FGFR3) signaling, leading to inhibition of mineralization through accumulation of the TNAP substrate pyrophosphate. Here, we report that the pyrophosphate concentration is increased in Hyp bones, and that Tnap expression is decreased in Hyp-derived osteocyte-like cells but not in Hyp-derived osteoblasts ex vivo and in vitro. In situ mRNA expression profiling in bone cryosections revealed a ~70-fold up-regulation of Fgfr3 mRNA in osteocytes versus osteoblasts of Hyp mice. In addition, we show that blocking of increased Fgf23-FGFR3 signaling with anti-Fgf23 antibodies or an FGFR3 inhibitor partially restored the suppression of Tnap expression, phosphate production, and mineralization, and decreased pyrophosphate concentration in Hyp-derived osteocyte-like cells in vitro. In vivo, bone-specific deletion of Fgf23 in Hyp mice rescued the suppressed TNAP activity in osteocytes of Hyp mice. Moreover, treatment of wild-type osteoblasts or mice with recombinant FGF23 suppressed Tnap mRNA expression and increased pyrophosphate concentrations in the culture medium and in bone, respectively. In conclusion, we found that the cell autonomous increase in Fgf23 secretion in Hyp osteocytes drives the accumulation of pyrophosphate through auto-/paracrine suppression of TNAP. Hence, we have identified a novel mechanism contributing to the mineralization defect in Hyp mice.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Francis F, Hennig S, Korn B, Reinhardt R, de Jong P, Poustka A, et al. A gene (PEX) with homologies to endopeptidases is mutated in patients with X-linked hypophosphatemic rickets. Nat Genet. 1995;11: 130–6. - PubMed
-
- Rowe PS, Oudet CL, Francis F, Sinding C, Pannetier S, Econs MJ, et al. Distribution of mutations in the PEX gene in families with X-linked hypophosphataemic rickets (HYP). Hum Mol Genet. 1997;6: 539–49. - PubMed
-
- Du L, Desbarats M, Viel J, Glorieux FH, Cawthorn C, Ecarot B. cDNA cloning of the murine Pex gene implicated in X-linked hypophosphatemia and evidence for expression in bone. Genomics. 1996;36: 22–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

