Functional Stability of the Human Kappa Opioid Receptor Reconstituted in Nanodiscs Revealed by a Time-Resolved Scintillation Proximity Assay
- PMID: 27035823
- PMCID: PMC4817975
- DOI: 10.1371/journal.pone.0150658
Functional Stability of the Human Kappa Opioid Receptor Reconstituted in Nanodiscs Revealed by a Time-Resolved Scintillation Proximity Assay
Abstract
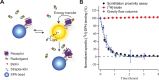
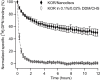
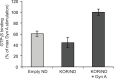
Long-term functional stability of isolated membrane proteins is crucial for many in vitro applications used to elucidate molecular mechanisms, and used for drug screening platforms in modern pharmaceutical industry. Compared to soluble proteins, the understanding at the molecular level of membrane proteins remains a challenge. This is partly due to the difficulty to isolate and simultaneously maintain their structural and functional stability, because of their hydrophobic nature. Here we show, how scintillation proximity assay can be used to analyze time-resolved high-affinity ligand binding to membrane proteins solubilized in various environments. The assay was used to establish conditions that preserved the biological function of isolated human kappa opioid receptor. In detergent solution the receptor lost high-affinity ligand binding to a radiolabelled ligand within minutes at room temperature. After reconstitution in Nanodiscs made of phospholipid bilayer the half-life of high-affinity ligand binding to the majority of receptors increased 70-fold compared to detergent solubilized receptors--a level of stability that is appropriate for further downstream applications. Time-resolved scintillation proximity assay has the potential to screen numerous conditions in parallel to obtain high levels of stable and active membrane proteins, which are intrinsically unstable in detergent solution, and with minimum material consumption.
Conflict of interest statement
Figures


Similar articles
-
Real-time monitoring of binding events on a thermostabilized human A2A receptor embedded in a lipid bilayer by surface plasmon resonance.Biochim Biophys Acta. 2015 May;1848(5):1224-33. doi: 10.1016/j.bbamem.2015.02.014. Epub 2015 Feb 25. Biochim Biophys Acta. 2015. PMID: 25725488
-
Reconstitution of Detergent-Solubilized Membrane Proteins into Proteoliposomes and Nanodiscs for Functional and Structural Studies.Methods Mol Biol. 2021;2302:21-35. doi: 10.1007/978-1-0716-1394-8_2. Methods Mol Biol. 2021. PMID: 33877620
-
Radioligand binding to nanodisc-reconstituted membrane transporters assessed by the scintillation proximity assay.Biochemistry. 2014 Jan 14;53(1):4-6. doi: 10.1021/bi401412e. Epub 2013 Dec 30. Biochemistry. 2014. PMID: 24344975 Free PMC article.
-
The nanodisc: a novel tool for membrane protein studies.Biol Chem. 2009 Aug;390(8):805-14. doi: 10.1515/BC.2009.091. Biol Chem. 2009. PMID: 19453280 Review.
-
Opioid receptor types and subtypes: the delta receptor as a model.Annu Rev Pharmacol Toxicol. 1996;36:379-401. doi: 10.1146/annurev.pa.36.040196.002115. Annu Rev Pharmacol Toxicol. 1996. PMID: 8725395 Review.
Cited by
-
Rational antibody design for undruggable targets using kinetically controlled biomolecular probes.Sci Adv. 2021 Apr 16;7(16):eabe6397. doi: 10.1126/sciadv.abe6397. Print 2021 Apr. Sci Adv. 2021. PMID: 33863724 Free PMC article.
-
Label-free, real-time monitoring of membrane binding events at zeptomolar concentrations using frequency-locked optical microresonators.Nat Commun. 2024 Aug 28;15(1):7445. doi: 10.1038/s41467-024-51320-x. Nat Commun. 2024. PMID: 39198447 Free PMC article.
-
Label-free, real-time monitoring of membrane binding events at zeptomolar concentrations using frequency-locked optical microresonators.bioRxiv [Preprint]. 2023 Sep 22:2023.09.20.558657. doi: 10.1101/2023.09.20.558657. bioRxiv. 2023. Update in: Nat Commun. 2024 Aug 28;15(1):7445. doi: 10.1038/s41467-024-51320-x. PMID: 37786702 Free PMC article. Updated. Preprint.
References
-
- Yıldırım MA, Goh K-I, Cusick ME, Barabási A-L, Vidal M. Drug—target network. Nat Biotechnol. 2007;25: 1119–1126. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

