Translational informatics approach for identifying the functional molecular communicators linking coronary artery disease, infection and inflammation
- PMID: 27035874
- PMCID: PMC4838147
- DOI: 10.3892/mmr.2016.5013
Translational informatics approach for identifying the functional molecular communicators linking coronary artery disease, infection and inflammation
Abstract
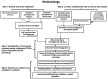
Translational informatics approaches are required for the integration of diverse and accumulating data to enable the administration of effective translational medicine specifically in complex diseases such as coronary artery disease (CAD). In the current study, a novel approach for elucidating the association between infection, inflammation and CAD was used. Genes for CAD were collected from the CAD‑gene database and those for infection and inflammation were collected from the UniProt database. The cytomegalovirus (CMV)‑induced genes were identified from the literature and the CAD‑associated clinical phenotypes were obtained from the Unified Medical Language System. A total of 55 gene ontologies (GO) termed functional communicator ontologies were identified in the gene sets linking clinical phenotypes in the diseasome network. The network topology analysis suggested that important functions including viral entry, cell adhesion, apoptosis, inflammatory and immune responses networked with clinical phenotypes. Microarray data was extracted from the Gene Expression Omnibus (dataset: GSE48060) for highly networked disease myocardial infarction. Further analysis of differentially expressed genes and their GO terms suggested that CMV infection may trigger a xenobiotic response, oxidative stress, inflammation and immune modulation. Notably, the current study identified γ‑glutamyl transferase (GGT)‑5 as a potential biomarker with an odds ratio of 1.947, which increased to 2.561 following the addition of CMV and CMV‑neutralizing antibody (CMV‑NA) titers. The C‑statistics increased from 0.530 for conventional risk factors (CRFs) to 0.711 for GGT in combination with the above mentioned infections and CRFs. Therefore, the translational informatics approach used in the current study identified a potential molecular mechanism for CMV infection in CAD, and a potential biomarker for risk prediction.
Figures

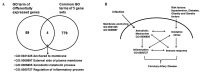
Similar articles
-
Integrative gene ontology and network analysis of coronary artery disease associated genes suggests potential role of ErbB pathway gene EGFR.Mol Med Rep. 2018 Mar;17(3):4253-4264. doi: 10.3892/mmr.2018.8393. Epub 2018 Jan 8. Mol Med Rep. 2018. PMID: 29328373 Free PMC article.
-
Integrated microRNA‑gene analysis of coronary artery disease based on miRNA and gene expression profiles.Mol Med Rep. 2016 Apr;13(4):3063-73. doi: 10.3892/mmr.2016.4936. Epub 2016 Feb 23. Mol Med Rep. 2016. PMID: 26936111 Free PMC article.
-
Role of Chlamydia pneumoniae, helicobacter pylori and cytomegalovirus in coronary artery disease.Pak J Pharm Sci. 2011 Apr;24(2):95-101. Pak J Pharm Sci. 2011. PMID: 21454155
-
Inflammation and coronary artery disease: insights from genetic studies.Can J Cardiol. 2012 Nov-Dec;28(6):662-6. doi: 10.1016/j.cjca.2012.05.014. Epub 2012 Aug 15. Can J Cardiol. 2012. PMID: 22902153 Review.
-
Evidence of systemic inflammation and estimation of coronary artery disease risk: a population perspective.Am J Med. 2008 Oct;121(10 Suppl 1):S15-20. doi: 10.1016/j.amjmed.2008.06.012. Am J Med. 2008. PMID: 18926165 Review.
Cited by
-
Integrative transcriptomic, proteomic, and machine learning approach to identifying feature genes of atrial fibrillation using atrial samples from patients with valvular heart disease.BMC Cardiovasc Disord. 2021 Jan 28;21(1):52. doi: 10.1186/s12872-020-01819-0. BMC Cardiovasc Disord. 2021. PMID: 33509101 Free PMC article.
-
In-Cardiome: integrated knowledgebase for coronary artery disease enabling translational research.Database (Oxford). 2017 Jan 1;2017:bax077. doi: 10.1093/database/bax077. Database (Oxford). 2017. PMID: 29220465 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

