Discrete gene replication events drive coupling between the cell cycle and circadian clocks
- PMID: 27035936
- PMCID: PMC4839394
- DOI: 10.1073/pnas.1507291113
Discrete gene replication events drive coupling between the cell cycle and circadian clocks
Abstract
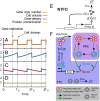
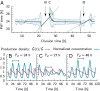
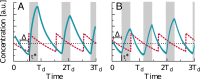
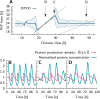
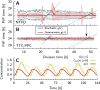
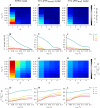
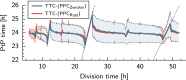
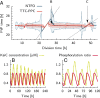
Many organisms possess both a cell cycle to control DNA replication and a circadian clock to anticipate changes between day and night. In some cases, these two rhythmic systems are known to be coupled by specific, cross-regulatory interactions. Here, we use mathematical modeling to show that, additionally, the cell cycle generically influences circadian clocks in a nonspecific fashion: The regular, discrete jumps in gene-copy number arising from DNA replication during the cell cycle cause a periodic driving of the circadian clock, which can dramatically alter its behavior and impair its function. A clock built on negative transcriptional feedback either phase-locks to the cell cycle, so that the clock period tracks the cell division time, or exhibits erratic behavior. We argue that the cyanobacterium Synechococcus elongatus has evolved two features that protect its clock from such disturbances, both of which are needed to fully insulate it from the cell cycle and give it its observed robustness: a phosphorylation-based protein modification oscillator, together with its accompanying push-pull read-out circuit that responds primarily to the ratios of different phosphoform concentrations, makes the clock less susceptible to perturbations in protein synthesis; the presence of multiple, asynchronously replicating copies of the same chromosome diminishes the effect of replicating any single copy of a gene.
Keywords: Kai; cell cycle; circadian rhythms; oscillations; simulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The circadian clock ensures successful DNA replication in cyanobacteria.Proc Natl Acad Sci U S A. 2021 May 18;118(20):e2022516118. doi: 10.1073/pnas.2022516118. Proc Natl Acad Sci U S A. 2021. PMID: 33972427 Free PMC article.
-
Robust circadian clocks from coupled protein-modification and transcription-translation cycles.Proc Natl Acad Sci U S A. 2010 Dec 28;107(52):22540-5. doi: 10.1073/pnas.1007613107. Epub 2010 Dec 13. Proc Natl Acad Sci U S A. 2010. PMID: 21149676 Free PMC article.
-
A mathematical model for the Kai-protein-based chemical oscillator and clock gene expression rhythms in cyanobacteria.J Biol Rhythms. 2007 Feb;22(1):69-80. doi: 10.1177/0748730406295749. J Biol Rhythms. 2007. PMID: 17229926
-
Molecular Links between the Circadian Clock and the Cell Cycle.J Mol Biol. 2020 May 29;432(12):3515-3524. doi: 10.1016/j.jmb.2020.04.003. Epub 2020 Apr 15. J Mol Biol. 2020. PMID: 32304699 Review.
-
Diversity of KaiC-based timing systems in marine Cyanobacteria.Mar Genomics. 2014 Apr;14:3-16. doi: 10.1016/j.margen.2013.12.006. Epub 2014 Jan 3. Mar Genomics. 2014. PMID: 24388874 Review.
Cited by
-
E-box binding transcription factors in cancer.Front Oncol. 2023 Aug 3;13:1223208. doi: 10.3389/fonc.2023.1223208. eCollection 2023. Front Oncol. 2023. PMID: 37601651 Free PMC article. Review.
-
A thermodynamically consistent model of the post-translational Kai circadian clock.PLoS Comput Biol. 2017 Mar 15;13(3):e1005415. doi: 10.1371/journal.pcbi.1005415. eCollection 2017 Mar. PLoS Comput Biol. 2017. PMID: 28296888 Free PMC article.
-
Modeling-Based Investigation of the Effect of Noise in Cellular Systems.Front Mol Biosci. 2018 Apr 12;5:34. doi: 10.3389/fmolb.2018.00034. eCollection 2018. Front Mol Biosci. 2018. PMID: 29707543 Free PMC article. Review.
-
Computational modelling unravels the precise clockwork of cyanobacteria.Interface Focus. 2018 Dec 6;8(6):20180038. doi: 10.1098/rsfs.2018.0038. Epub 2018 Oct 19. Interface Focus. 2018. PMID: 30443335 Free PMC article. Review.
-
Synchronizing stochastic circadian oscillators in single cells of Neurospora crassa.Sci Rep. 2016 Oct 27;6:35828. doi: 10.1038/srep35828. Sci Rep. 2016. PMID: 27786253 Free PMC article.
References
-
- Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 2003;426(6967):680–683. - PubMed
-
- Soták M, Sumová A, Pácha J. Cross-talk between the circadian clock and the cell cycle in cancer. Ann Med. 2014;46(4):221–232. - PubMed
-
- Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302(5643):255–259. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

