Twist-open mechanism of DNA damage recognition by the Rad4/XPC nucleotide excision repair complex
- PMID: 27035942
- PMCID: PMC4843471
- DOI: 10.1073/pnas.1514666113
Twist-open mechanism of DNA damage recognition by the Rad4/XPC nucleotide excision repair complex
Abstract
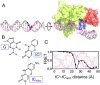
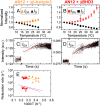
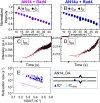
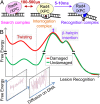
DNA damage repair starts with the recognition of damaged sites from predominantly normal DNA. In eukaryotes, diverse DNA lesions from environmental sources are recognized by the xeroderma pigmentosum C (XPC) nucleotide excision repair complex. Studies of Rad4 (radiation-sensitive 4; yeast XPC ortholog) showed that Rad4 "opens" up damaged DNA by inserting a β-hairpin into the duplex and flipping out two damage-containing nucleotide pairs. However, this DNA lesion "opening" is slow (˜5-10 ms) compared with typical submillisecond residence times per base pair site reported for various DNA-binding proteins during 1D diffusion on DNA. To address the mystery as to how Rad4 pauses to recognize lesions during diffusional search, we examine conformational dynamics along the lesion recognition trajectory using temperature-jump spectroscopy. Besides identifying the ˜10-ms step as the rate-limiting bottleneck towards opening specific DNA site, we uncover an earlier ˜100- to 500-μs step that we assign to nonspecific deformation (unwinding/"twisting") of DNA by Rad4. The β-hairpin is not required to unwind or to overcome the bottleneck but is essential for full nucleotide-flipping. We propose that Rad4 recognizes lesions in a step-wise "twist-open" mechanism, in which preliminary twisting represents Rad4 interconverting between search and interrogation modes. Through such conformational switches compatible with rapid diffusion on DNA, Rad4 may stall preferentially at a lesion site, offering time to open DNA. This study represents the first direct observation, to our knowledge, of dynamical DNA distortions during search/interrogation beyond base pair breathing. Submillisecond interrogation with preferential stalling at cognate sites may be common to various DNA-binding proteins.
Keywords: DNA damage recognition; DNA unwinding dynamics; temperature-jump perturbation; time-resolved fluorescence spectroscopy; xeroderma pigmentosum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Enhanced spontaneous DNA twisting/bending fluctuations unveiled by fluorescence lifetime distributions promote mismatch recognition by the Rad4 nucleotide excision repair complex.Nucleic Acids Res. 2018 Feb 16;46(3):1240-1255. doi: 10.1093/nar/gkx1216. Nucleic Acids Res. 2018. PMID: 29267981 Free PMC article.
-
Sequence specificity, energetics and mechanism of mismatch recognition by DNA damage sensing protein Rad4/XPC.Nucleic Acids Res. 2020 Mar 18;48(5):2246-2257. doi: 10.1093/nar/gkaa078. Nucleic Acids Res. 2020. PMID: 32047903 Free PMC article.
-
Kinetic gating mechanism of DNA damage recognition by Rad4/XPC.Nat Commun. 2015 Jan 6;6:5849. doi: 10.1038/ncomms6849. Nat Commun. 2015. PMID: 25562780 Free PMC article.
-
XPC: Going where no DNA damage sensor has gone before.DNA Repair (Amst). 2015 Dec;36:19-27. doi: 10.1016/j.dnarep.2015.09.004. Epub 2015 Sep 9. DNA Repair (Amst). 2015. PMID: 26422135 Free PMC article. Review.
-
Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair.Mol Carcinog. 2003 Sep;38(1):1-13. doi: 10.1002/mc.10143. Mol Carcinog. 2003. PMID: 12949838 Review.
Cited by
-
Melting temperature measurement and mesoscopic evaluation of single, double and triple DNA mismatches.Chem Sci. 2020 Jul 23;11(31):8273-8287. doi: 10.1039/d0sc01700k. Chem Sci. 2020. PMID: 34094181 Free PMC article.
-
Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.Nat Commun. 2021 Jun 7;12(1):3338. doi: 10.1038/s41467-021-23684-x. Nat Commun. 2021. PMID: 34099686 Free PMC article.
-
Nucleotide Excision Repair: Insights into Canonical and Emerging Functions of the Transcription/DNA Repair Factor TFIIH.Genes (Basel). 2025 Feb 19;16(2):231. doi: 10.3390/genes16020231. Genes (Basel). 2025. PMID: 40004560 Free PMC article. Review.
-
Enhanced spontaneous DNA twisting/bending fluctuations unveiled by fluorescence lifetime distributions promote mismatch recognition by the Rad4 nucleotide excision repair complex.Nucleic Acids Res. 2018 Feb 16;46(3):1240-1255. doi: 10.1093/nar/gkx1216. Nucleic Acids Res. 2018. PMID: 29267981 Free PMC article.
-
Structural modeling and analyses of genetic variations in the human XPC nucleotide excision repair protein.J Biomol Struct Dyn. 2023;41(23):13535-13562. doi: 10.1080/07391102.2023.2177349. Epub 2023 Mar 8. J Biomol Struct Dyn. 2023. PMID: 36890638 Free PMC article.
References
-
- Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970;53(3):401–417. - PubMed
-
- Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor--operator interaction: Kinetic measurements and conclusions. Biochemistry. 1981;20(24):6961–6977. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

