Modeling and analysis of collective cell migration in an in vivo three-dimensional environment
- PMID: 27035964
- PMCID: PMC4839456
- DOI: 10.1073/pnas.1522656113
Modeling and analysis of collective cell migration in an in vivo three-dimensional environment
Abstract
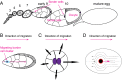
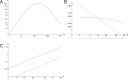
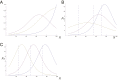
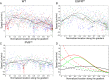
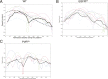
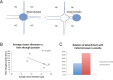
A long-standing question in collective cell migration has been what might be the relative advantage of forming a cluster over migrating individually. Does an increase in the size of a collectively migrating group of cells enable them to sample the chemical gradient over a greater distance because the difference between front and rear of a cluster would be greater than for single cells? We combined theoretical modeling with experiments to study collective migration of the border cells in-between nurse cells in the Drosophila egg chamber. We discovered that cluster size is positively correlated with migration speed, up to a particular point above which speed plummets. This may be due to the effect of viscous drag from surrounding nurse cells together with confinement of all of the cells within a stiff extracellular matrix. The model predicts no relationship between cluster size and velocity for cells moving on a flat surface, in contrast to movement within a 3D environment. Our analyses also suggest that the overall chemoattractant profile in the egg chamber is likely to be exponential, with the highest concentration in the oocyte. These findings provide insights into collective chemotaxis by combining theoretical modeling with experimentation.
Keywords: cell migration; chemotaxis; theoretical modeling; three-dimensional.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Parent CA, Devreotes PN. A cell’s sense of direction. Science. 1999;284(5415):765–770. - PubMed
-
- Devreotes PN, Zigmond SH. Chemotaxis in eukaryotic cells: A focus on leukocytes and Dictyostelium. Annu Rev Cell Biol. 1988;4:649–686. - PubMed
-
- Yu SR, et al. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature. 2009;461(7263):533–536. - PubMed
-
- Crick F. Diffusion in embryogenesis. Nature. 1970;225(5231):420–422. - PubMed
-
- Moissoglu K, Majumdar R, Parent CA. Cell migration: Sinking in a gradient. Curr Biol. 2014;24(1):R23–R25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

