Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria
- PMID: 27035970
- PMCID: PMC4839414
- DOI: 10.1073/pnas.1523926113
Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria
Abstract
Selective autophagy of damaged mitochondria requires autophagy receptors optineurin (OPTN), NDP52 (CALCOCO2), TAX1BP1, and p62 (SQSTM1) linking ubiquitinated cargo to autophagic membranes. By using quantitative proteomics, we show that Tank-binding kinase 1 (TBK1) phosphorylates all four receptors on several autophagy-relevant sites, including the ubiquitin- and LC3-binding domains of OPTN and p62/SQSTM1 as well as the SKICH domains of NDP52 and TAX1BP1. Constitutive interaction of TBK1 with OPTN and the ability of OPTN to bind to ubiquitin chains are essential for TBK1 recruitment and kinase activation on mitochondria. TBK1 in turn phosphorylates OPTN's UBAN domain at S473, thereby expanding the binding capacity of OPTN to diverse Ub chains. In combination with phosphorylation of S177 and S513, this posttranslational modification promotes recruitment and retention of OPTN/TBK1 on ubiquitinated, damaged mitochondria. Moreover, phosphorylation of OPTN on S473 enables binding to pS65 Ub chains and is also implicated in PINK1-driven and Parkin-independent mitophagy. Thus, TBK1-mediated phosphorylation of autophagy receptors creates a signal amplification loop operating in selective autophagy of damaged mitochondria.
Keywords: OPTN; TBK1; mitophagy; phosphorylation; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

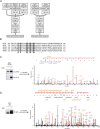
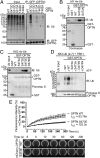






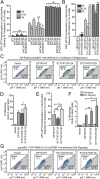


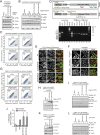
Comment in
-
The TBK1-OPTN Axis Mediates Crosstalk Between Mitophagy and the Innate Immune Response: A Potential Therapeutic Target for Neurodegenerative Diseases.Neurosci Bull. 2017 Jun;33(3):354-356. doi: 10.1007/s12264-017-0116-3. Epub 2017 Mar 7. Neurosci Bull. 2017. PMID: 28271436 Free PMC article. No abstract available.
Similar articles
-
The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy.Mol Cell. 2015 Oct 1;60(1):7-20. doi: 10.1016/j.molcel.2015.08.016. Epub 2015 Sep 10. Mol Cell. 2015. PMID: 26365381 Free PMC article.
-
Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy.Proc Natl Acad Sci U S A. 2016 Jun 14;113(24):E3349-58. doi: 10.1073/pnas.1523810113. Epub 2016 May 31. Proc Natl Acad Sci U S A. 2016. PMID: 27247382 Free PMC article.
-
Structural insights into the ubiquitin recognition by OPTN (optineurin) and its regulation by TBK1-mediated phosphorylation.Autophagy. 2018;14(1):66-79. doi: 10.1080/15548627.2017.1391970. Epub 2018 Feb 2. Autophagy. 2018. PMID: 29394115 Free PMC article.
-
Two different axes CALCOCO2-RB1CC1 and OPTN-ATG9A initiate PRKN-mediated mitophagy.Autophagy. 2020 Nov;16(11):2105-2107. doi: 10.1080/15548627.2020.1815457. Epub 2020 Sep 7. Autophagy. 2020. PMID: 32892694 Free PMC article. Review.
-
[Research progress on the role of TANK-binding kinase 1 in PINK1/Parkin-dependent and -independent mitophagy].Sheng Li Xue Bao. 2024 Feb 25;76(1):161-172. Sheng Li Xue Bao. 2024. PMID: 38444141 Review. Chinese.
Cited by
-
Cerebellar Kv3.3 potassium channels activate TANK-binding kinase 1 to regulate trafficking of the cell survival protein Hax-1.Nat Commun. 2021 Mar 19;12(1):1731. doi: 10.1038/s41467-021-22003-8. Nat Commun. 2021. PMID: 33741962 Free PMC article.
-
Role of autophagy in tumor response to radiation: Implications for improving radiotherapy.Front Oncol. 2022 Sep 12;12:957373. doi: 10.3389/fonc.2022.957373. eCollection 2022. Front Oncol. 2022. PMID: 36172166 Free PMC article. Review.
-
IKKε and TBK1 prevent RIPK1 dependent and independent inflammation.Nat Commun. 2024 Jan 2;15(1):130. doi: 10.1038/s41467-023-44372-y. Nat Commun. 2024. PMID: 38167258 Free PMC article.
-
PINK1/TAX1BP1-directed mitophagy attenuates vascular endothelial injury induced by copper oxide nanoparticles.J Nanobiotechnology. 2022 Mar 19;20(1):149. doi: 10.1186/s12951-022-01338-4. J Nanobiotechnology. 2022. PMID: 35305662 Free PMC article.
-
NDP52 interacts with mitochondrial RNA poly(A) polymerase to promote mitophagy.EMBO Rep. 2018 Dec;19(12):e46363. doi: 10.15252/embr.201846363. Epub 2018 Oct 11. EMBO Rep. 2018. PMID: 30309841 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

