Measuring and modeling diffuse scattering in protein X-ray crystallography
- PMID: 27035972
- PMCID: PMC4839442
- DOI: 10.1073/pnas.1524048113
Measuring and modeling diffuse scattering in protein X-ray crystallography
Abstract
X-ray diffraction has the potential to provide rich information about the structural dynamics of macromolecules. To realize this potential, both Bragg scattering, which is currently used to derive macromolecular structures, and diffuse scattering, which reports on correlations in charge density variations, must be measured. Until now, measurement of diffuse scattering from protein crystals has been scarce because of the extra effort of collecting diffuse data. Here, we present 3D measurements of diffuse intensity collected from crystals of the enzymes cyclophilin A and trypsin. The measurements were obtained from the same X-ray diffraction images as the Bragg data, using best practices for standard data collection. To model the underlying dynamics in a practical way that could be used during structure refinement, we tested translation-libration-screw (TLS), liquid-like motions (LLM), and coarse-grained normal-modes (NM) models of protein motions. The LLM model provides a global picture of motions and was refined against the diffuse data, whereas the TLS and NM models provide more detailed and distinct descriptions of atom displacements, and only used information from the Bragg data. Whereas different TLS groupings yielded similar Bragg intensities, they yielded different diffuse intensities, none of which agreed well with the data. In contrast, both the LLM and NM models agreed substantially with the diffuse data. These results demonstrate a realistic path to increase the number of diffuse datasets available to the wider biosciences community and indicate that dynamics-inspired NM structural models can simultaneously agree with both Bragg and diffuse scattering.
Keywords: diffuse scattering; liquid-like motions; normal modes; protein dynamics; structural biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
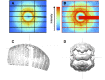
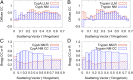

References
-
- Clarage JB, Phillips GN., Jr Analysis of diffuse scattering and relation to molecular motion. Methods Enzymol. 1997;277:407–432. - PubMed
-
- Keen DA, Goodwin AL. The crystallography of correlated disorder. Nature. 2015;521(7552):303–309. - PubMed
-
- Schomaker V, Trueblood KN. On the rigid-body motion of molecules in crystals. Acta Crystallogr B. 1968;24(1):63–76.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

